DNA damage and repair
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
46 Terms
why can DNA damage result in cell death?
transcription and replication can be blocked, cell cycle stopped→
what do mutations arise from?
-errors replication of undamaged DNA
-errors replication of damaged DNA
-inaccurate repair damaged DNA
what are most mutations from tho?
inaccurate repair of damaged DNA
how much less do mutations form in sperm cells? (compared to somatic cells)
10x lower
what are the 2 ways DNA damage can occur?
spontaneously since DNA is chemical (e.g. oxidative), external agents (mutagens)
what are the 3 spontaneous ways DNA can be damaged?
chemical: deamination of cytosine, depurination
damage from reactive oxygen species (H2O2 & free radicals)
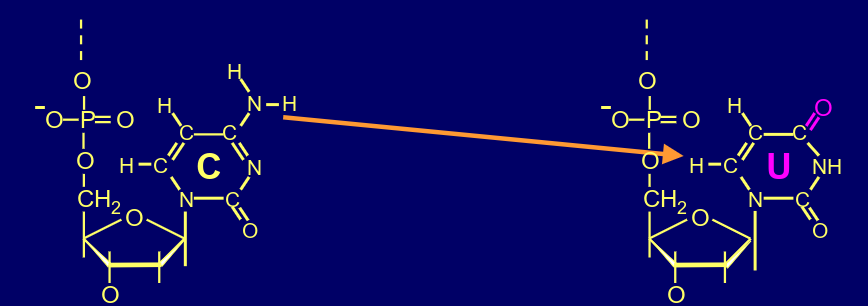
whats this?
deamination of cytosine
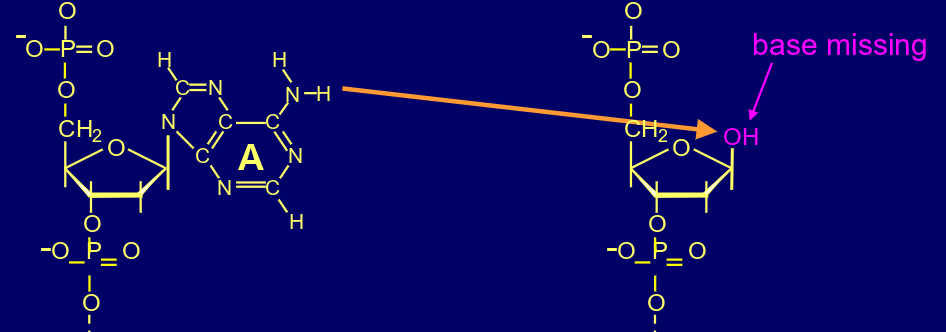
whats this?
depurination
what can the deamination of C cause?
C-G can switch to A-T pairing (if not repaired)
what can the depurination cause?
apurinic sites block transcription & can block replication
what are the 4 ways ROS interact with DNA?
chemical modification of bases (oxidise), formation of cyclopurines (structural distortion), interstrand cross-links, strand breaks
what are the 3 ways DNA damage can be induced?
ionising radiation (x-rays), ultraviolet (UV) radiation, chemicals (e.g. polycyclic aromatic hydrocarbons)
whats one of the most main forms of radiation?
radon (more or less exposure depending on where you live)
how does ionising radiation fuck up your dna?
creates radicals
exposure of UV to DNA causes?
cyclobutane pyrimidine dimers (CPDs), (covalent bonds between adjacent pyrimidines), formation of 6-4 photoproducts
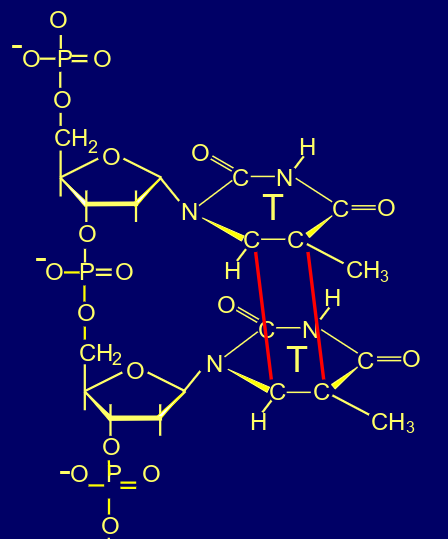
whats this
cyclobutane pyrimidine dimer
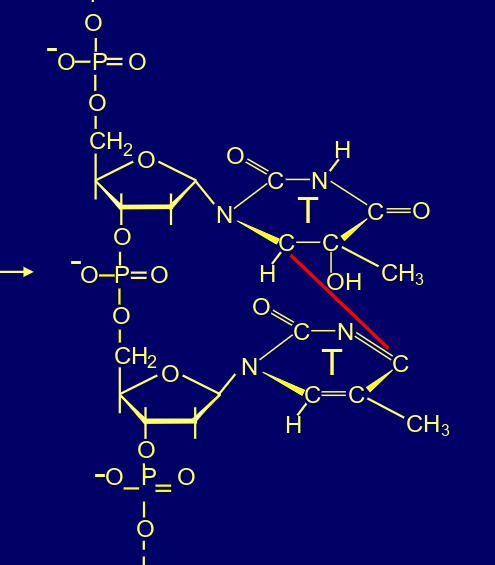
whats this?
6-4 photoproduct
why is it bad photoproducts cause structural distortions in DNA?
prevents normal passage of DNA polymerase, and rna polymerase
what kind of uv creates most to least amount of CPDs?
UVC (100x more than) UVB (1000x more than) UVA
what are the usually good DNA repair mechanisms?
base excision repair, nucleotide excision repair, mismatch repair, homology directed repair
what are the error-prone DNA repair mechanisms?
non-homologous end joining, translesion sythesis (bypass damage)
how is cytosine deamination repaired?
base excision repair
how is cytosine depurination repaired?
base excision repair
how is base damage from ROS repaired?
base excision repair
how is interstrand crosslink repaired?
interstrand crosslink repair
how is interstrand crosslink caused?
some chemotherapy drugs
how is cyclobutane pyrimidine dimer repaired?
nucleotide excision repair
how is adduct repaired?
nucleotide excision repair
how is an adduct to DNA caused?
chemicals including PAHs
how is a double stranded break repaired?
homology-directed repair, non-homologous end joining
how does base excision repair work?
damage specific glycosylase enzymes slide along DNA , recognise and excise damaged bases → creates apurinic and apyrimidic sites
apurinic/ apyrimidic site endonuclease cleaves phosphodiester bond 5’ to the AP site
in mammals -lyase activity of beta DNA polymerase may remove the deoxyribose and phosphate previously linked to damaged base
DNA polymerase b replaces missing nucleotide, DNA ligase seals the gap
how does nucleotide excision repair work?
recognition of damage and unwinding → cleavage at sites flanking damage → DNA polymerase synthesise new DNA → DNA ligase seals gaps
what are the 2 types of nucleotide excision repair?
template-strand and global genome
how does GG-NER recognise dsmage?
the structural distortion detected by protein XPC
what does XPC recruit to help it recognise CPDs
UV-DDB
how does TS-NER recognise damage?
RNA polymerase stalls at damaged site, proteins CSA and CSB are recruited, RNA polymerase backtracks, marking site and allowing NER machinery access
what happens next in nucleotide excision repair after site recognition?
helicase activity of TFIIH unwinds DNA near damage site,
damaged strand cleaved on both ends by XPG and XPF,
release of fragment containing damaged site, synthesis of DNA to fill gap,
synthesis of missing phosphodiester bond by DNA ligase
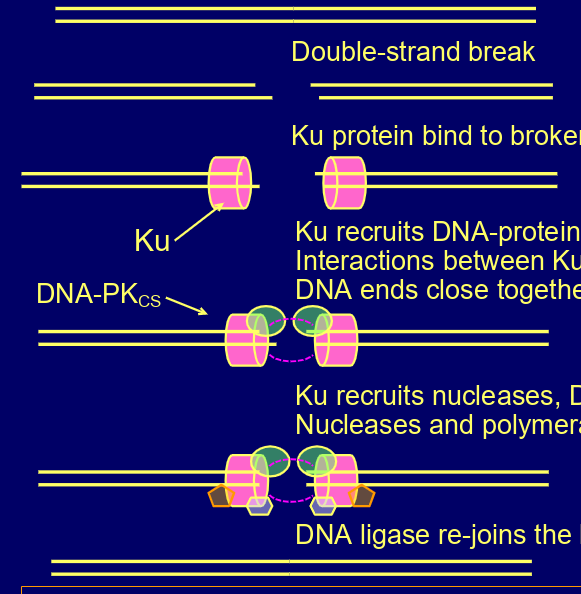
how does non-homologous end joining work?
ku protein bind to both ends, recruits DNA-protein kinase catalytic subunit- interactions keep them close,
ku recruits nucleases,
DNA polymerases & DNA ligases,
DNA ligases re-joins the broken ends
what are the consequences of DNA damage
-mutation
-ageing
what is the evidence DNA damage causes ageing?
mutations and DNA damage accumulate with age
all inherited disorders that lead to premature ageing come from mutations in genes involved in DNA repair and maintenance
cancer patients cured w chemotherapy that damages DNA show signs of premature ageing
what causes xeroderma pigmentosum?
due to mutations that compromise global genome- NER
what causes Cockayne syndrome?
mutations that compromise transcription coupled NER
what are the symptoms of xerodermal pigmentosum?
-heightened sensitivity of skin and eyes to sunlight
-increased risk of skin cancer
-neurological degeneration
what are the symptoms of Cockayne syndrome?
-extreme sensitivity of skin to sunlight
-progressive neurological degradation
-premature ageing (death of non-proliferating cells)
what is the mechanism of cockayne syndrome?
-RNA polymerase stalls at damaged site
-without functional CSB or CSA, RNA polymerase II remains stalled on the gene and fails to backtrack, making damaged region inaccessible to all repair mechanisms