Week 4 - Non-coding RNA & Hallmarks of Cancer
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
46 Terms
Coding genes percentage and info
1% of genome, 20,000 code for DNA and most are the same in other species.
What does noncoding DNA do?
Goes against central DOGMA (DNA → transcription → mRNA → translation → protein)
transcribe into noncoding RNA
The fine-tuning gene expression in uniquely human
(noncoding RNA = regulatory)
Regulatory RNAs
RNA can encode without being translated into protein
Natural forms:
microRNAs
long Noncoding RNAs
Long double stranded RNA (pathological)
Short interfering RNA (synthetic)
microRNAs
small non-protein coding RNAs
21-22 nucleotides long
double stranded
Where are genes for microRNAs found?
Found throughout genome, often in introns or intergenic
microRNA function (2 functions)
1. Degradation: miRNA matches sequence of target mRNA perfectly = target is degraded = no protein
2. Block translation: miRNA matches sequence of target mRNA imperfectly = inhibited from translation = less protein
[number 2 is more common]
miRNA can have how many targets?
Many
can have multiple functions and regulate various pathways.
Mir7
tumour suppressor miRNA
function = prevents invasion and migration in melanoma
downregulated in cancer
Mir200
miRNA family inhibit epithelial characteristics and promote mesenchymal traits.
function = suppresses ZEB1 and ZEB2 → which are in high conc. in mesenchymal cells and repress e-cadherin.
therefore Mir200 family help sustain high e-cadherin conc.
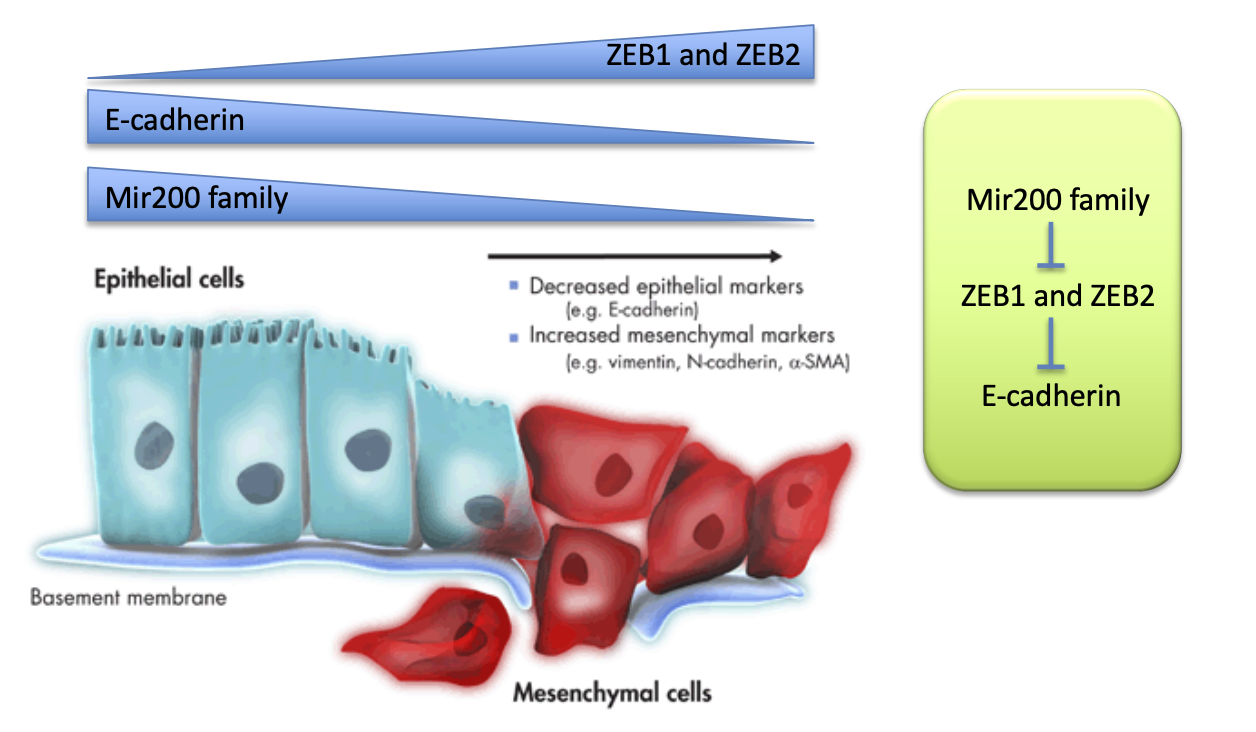
ZEB1/2 (related to Mir200)
Transcriptional repressors of e-cadherin = leads to low e-cadherin = promotes epithelial to mesenchymal transition
Mir200 suppresses it therefore maintaining e-cadherin levels.
Long noncoding RNAs
greater than 200 nucleotides
does no contain obvious protein coding potential
(a type of regulatory RNA)
each as a unique DNA, RNA & protein recognition motif
most never lease nucleus
Where are long noncoding RNAs found?
many locations:
- intergenic
- splice forms
- intronic
- antisense
LncRNA expression
lowly expressed
Specific to a tissue or development stage
The function of most are unknown
LncRNA mechanism of action (name the 4 types)
1. signals
2. decoys
3. guides
4. scaffolds
LncRNA are regulating genes are the transcriptional level
Archetype 1: Signal LncRNA
They mimic the combinatorial actions of transcriptions factors (or signalling pathways)
to form a response faster than making a new protein
LncRNA signal example → XIST
Role: X-inactivation
17kilobase lncRNA gene
found in X chromosome in placental mammals
In order for X-chromosomes in women to be expressed at the same rate as men, one must be silenced.
XIST function
Signal = showing which X is silenced
Guide = recruites epigenetic machinery to one X chromosome, to silence it
(essential for X-inactivation)
Archetype 2: Decoy LncRNA
sequester transcription factors away from chromatin
sequester protein factors into nuclear domains
can also be decoys for miRNA
Archetype 3: Guide LncRNA
recruit chromatin modifying enzymes to target genes either in
cis: near the site of the lncRNA production
or
trans: distant target genes
LncRNA guide example: HOTAIR
Found in HOX gene cluster
HOX genes are vital for patterning in the embryo
HOTAIR represses this by:
changing epigenetic state
Then inducing heterochromatin
Upregulated in many cancers (associated with metastasis)
Archetype 4: Scaffold LncRNA
Can bring together multiple proteins to form ribonucleoprotein complexes
complexes can alter gene expression
e.g. epigentic modification
Can also be structural to stabilise structures or signalling complexes
Scaffold LncRNA example: NEAT1
(decoy and scaffold)
23kb gene in mammals
Scaffold = structural scaffold for paraspeckles
Decoy = alters gene regulation by sequestering proteins and RNA
6 Hallmarks of cancer
1. Evade apoptosis
2. Self sufficient growth signals
3. Insensitive to anti-growth signals
4. Sustains angiogenesis
5. limitless replicative potential
6. tissue invasion and metastasis
Hallmark 1: Evading apoptosis (cancer)
mutations that:
- inactivate apoptotic sensors
- inactivate apoptotic effectors
Apoptotic Sensors
1. Cell surface receptors that bind survival/death factors
2. Intracellular sensors that monitor DNA damage
^ Signals tell mitochondria to release cytochrome C which is responsible for triggering apoptosis
Mutation in cancer cells needs to inactivate these sensors
Apoptotic Effectors
Capsases = enzymes that break down cell organelles
Inactivated by mutations in cancer cells
Apoptosis involves many effectors/capsases (drugs can kill the tumour by increasing levels of a different effector/s)
Hallmark 2: Self sufficiency in growth signals (3 ways cancer cells do it)
Tumour cells:
1. generate own growth signal
2. overexpress receptor for growth signals in a permanently activated forms
3. mutate downstream signalling molecules = permanent activation
Hallmark 3: Anti-growth signals in normal cells (overview)
in G1 phase:
- cells monitor environment
- can decide to put cell back to quiesccent or differentiated states
Anti-growth signals normal vs Cancer
Embryonic Myc-Max = keeps cells pluripotent
Mature cells Mad-Max = triggers differentiation
cancer reactivates Myc = keeps cells pluripotent
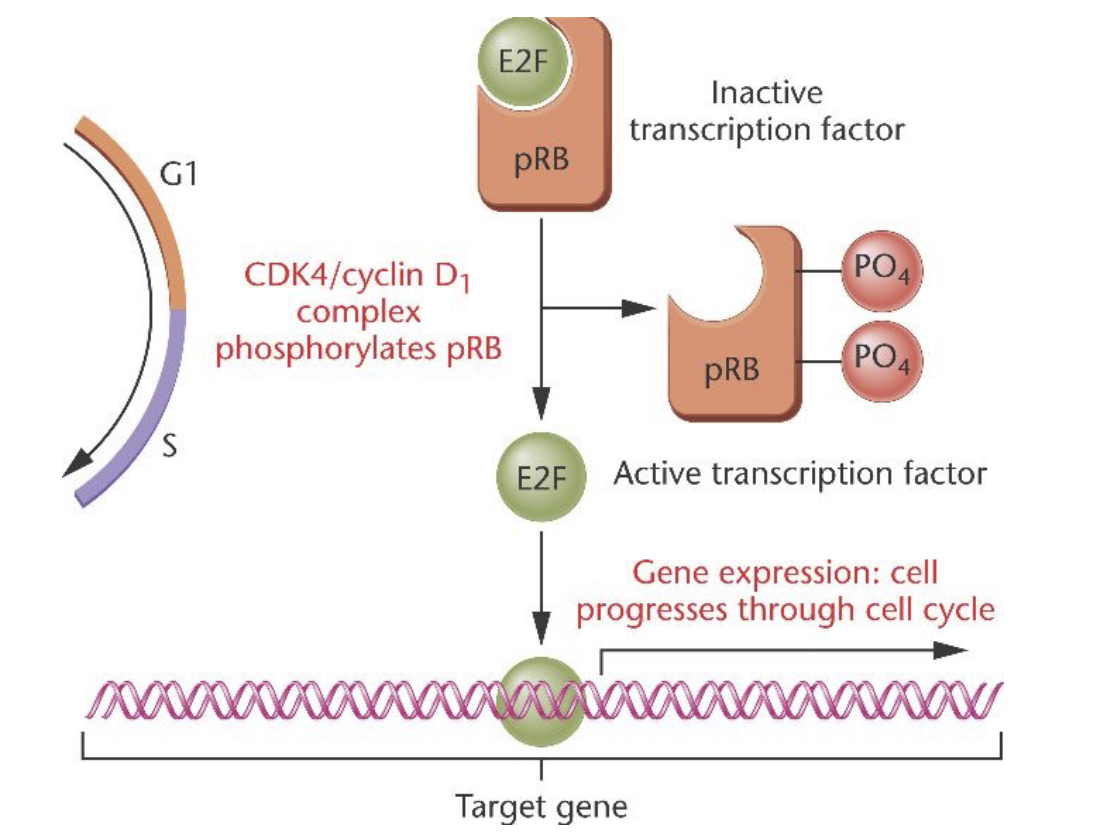
pRB/E2F pathway (relation to cancer)
directs G1 to S
Cancer:
- TGFbeta blocks phosphorylation of pRB, cancer cells must prevent TGFbeta from doing this
freeing up of E2F:
takes cell out of G1 phase into S phase
Hallmark 4: Sustained angiogenesis (cancer)
Early to mid stage step required by tumours
formation of macroscopic tumours
cells must be 100microns from blood vessel:
tumour redirects blood vessel growth towards it
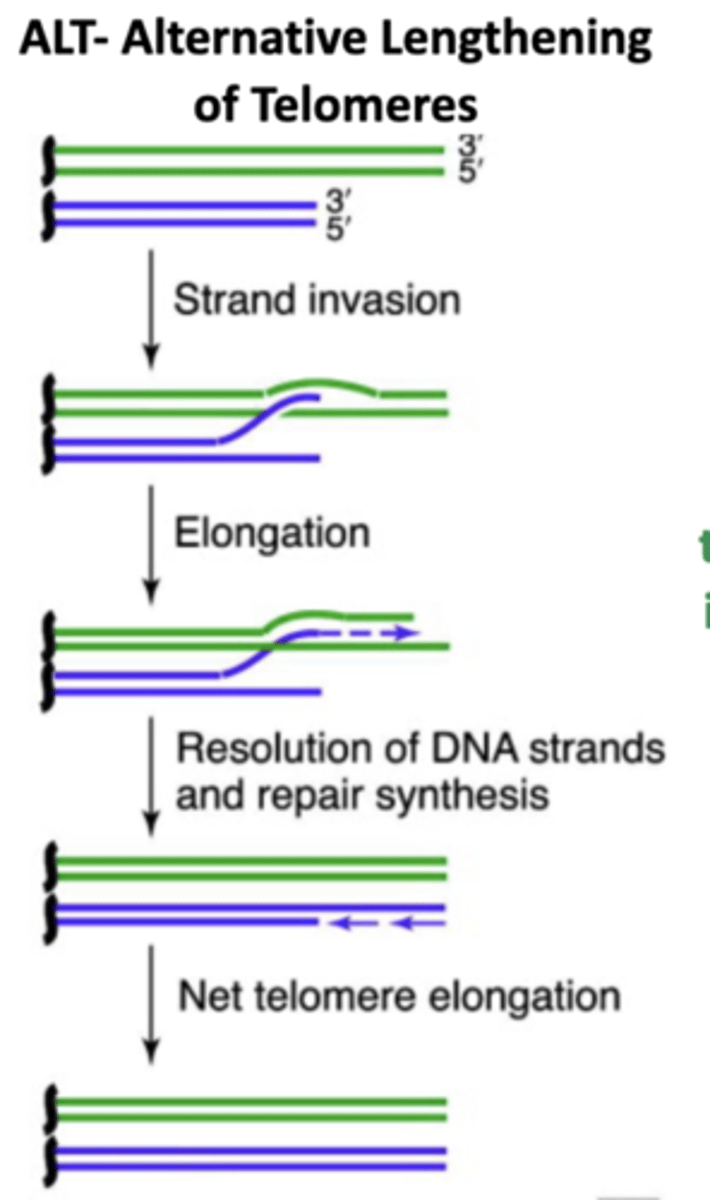
Hallmark 5: Limitless Replicative Potential (cancer)
Normal cells = enter senescence after a number of doublings
Cancer = continues replicating by maintaining Telomeres
How do cancer cells continue replicating?
Maintain Telomeres by:
- upregulate telomerase (80-90%)
- the ALT mechanism that uses homologous recombination based process (10-20%)
Hallmark 6: Metastasis and Tumour invasion (cancer) - Requires what 2 things?
changes to physical coupling of tumour cells to stroma (supporting tissue)
activation of extracellular proteases
These allow for the transition from epithelial to mesenchymal cells
E-cadherin in cancer
is lost in invasive epithelial cancers:
- tumours change the integrins they produce
- which effects what the cells contact with
microRNA200 family reduce in concentration.
Cancer and cell jucntions
Adherens junctions and focal adhesions must be dismantled to successful form mesenchymal cells
Enabling characteristics of Cancer (2)
1. Genome instability
2. Inflammation
Genome instability in cancer
Cancer = increase sensitivity to mutagenic agents → decreases efficiency of DNA repair
(cancer enabling characteristic)
Inflammation
Innate immune cells supply:
growth factors
ECM modifying enzymes
proangiogenic factors
Survival factors
release ROS
to help mutate nearby cells
Emerging hallmarks of Cancer (2)
1. Cellular energetics/metabolism
2. Avoiding immune system
Cellular Energetics/metabolism (in cancer)
Warburg effect:
- most cancer cells produce energy via glycolysis
- even in presence of oxygen → bypass need for ATP from mitochondria
- therefore cancer cells like glucose
Avoiding immune system
Not a universally recognised hallmark
sick people aren’t MORE prone to cancer
some cancers evade immune cells better than others
Multi Hit model of Carcinogenesis
Cancers develop over time as mutations accumulate
Cancers need all 6 of the hallmarks (acquires them through accumulating several mutations)
p53 tumour suppressor
Suppresses tumours
(if cancer cells are successful they evade it)
mutating P53 achieves insensitivity to anti-growth and evading apoptosis
hitting two hallmarks in one
Why do cancer drugs fail?
targetting one hallmark = may activate others
therefore combination therapies are needed
Limitation of Cancer Hallmark Model (2)
1. Some cancers rely heavily on one hallmark (each hallmark isnt equal)
2. First 5 hallmarks dont focus on Metastasis, which is the real problem with cancer