BIO 123 | Microbial Diversity I: Domain Bacteria & Archaea
1/248
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
249 Terms
Describe microbial diversity
Microorganisms account for majority of life on earth (>1 trillion in some estimates)
Microbes possess diverse array of metabolic activities, which allows them to colonize virtually every habitat
All 3 domains of life are dominated by microbial life present within
T/F: Only plants, animals, and fungi are macroorganisms.
TRUE
T/F: Microorganisms are dominant in all 3 domains of life.
TRUE
8 Challenges in Estimating Microbial Diversity
Strain concept
Changing phenotype
Functional role similarity
Horizontal gene transfer
Habitat specialization
Nonculturability
Defining a microorganism
Syphilis-causing bacterium
Treponema pallidum
Variety and variability of life
Diversity
T/F: Members of higher-level groups share fewer characteristics than those in lower-level groups.
TRUE
The more specific (the lower), the more characteristics they share.
As a general rule, _ is the smallest grouping of organisms possible.
species
_ is an exception to general rule that species is the smallest grouping of organisms possible
strain
When do you consider something a strain?
Small but permanent genetic difference
Antibiotic resistance (tricky bc they can get it thru HGT)
Need for a nutrient (thats not typically needed by species of that group)
Antigen presence
T/F: Unclassified bacteria is valuable bc it can contribute to overall similarities and differences that we don’t know of.
TRUE
T/F: Strains can be defined by a single nucleotide change, which may change the chromosomal structure of an organism.
TRUE
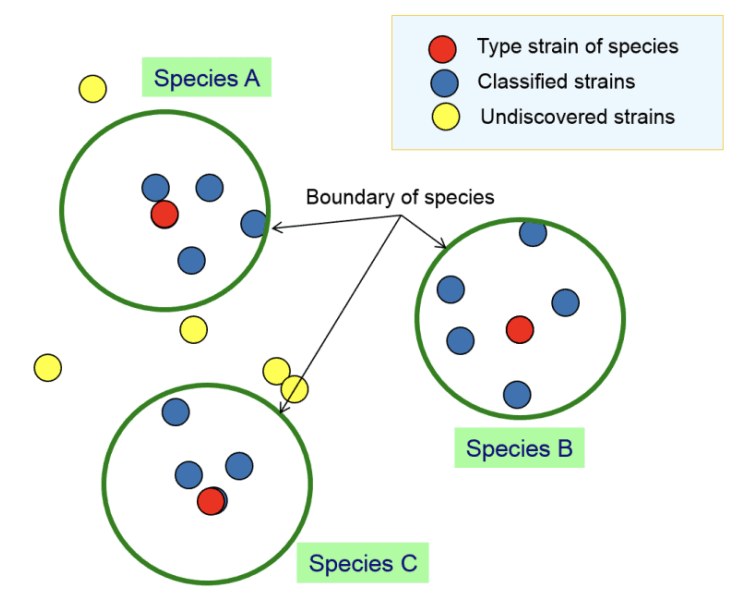
Explain
Type strain: type specimen, a standard or reference specimen to which all classified strains should be close in terms of properties.
Classified strains
Undiscovered strains: unclassified
If animals have subspecies, plants have tribes, bacteria have _
strains
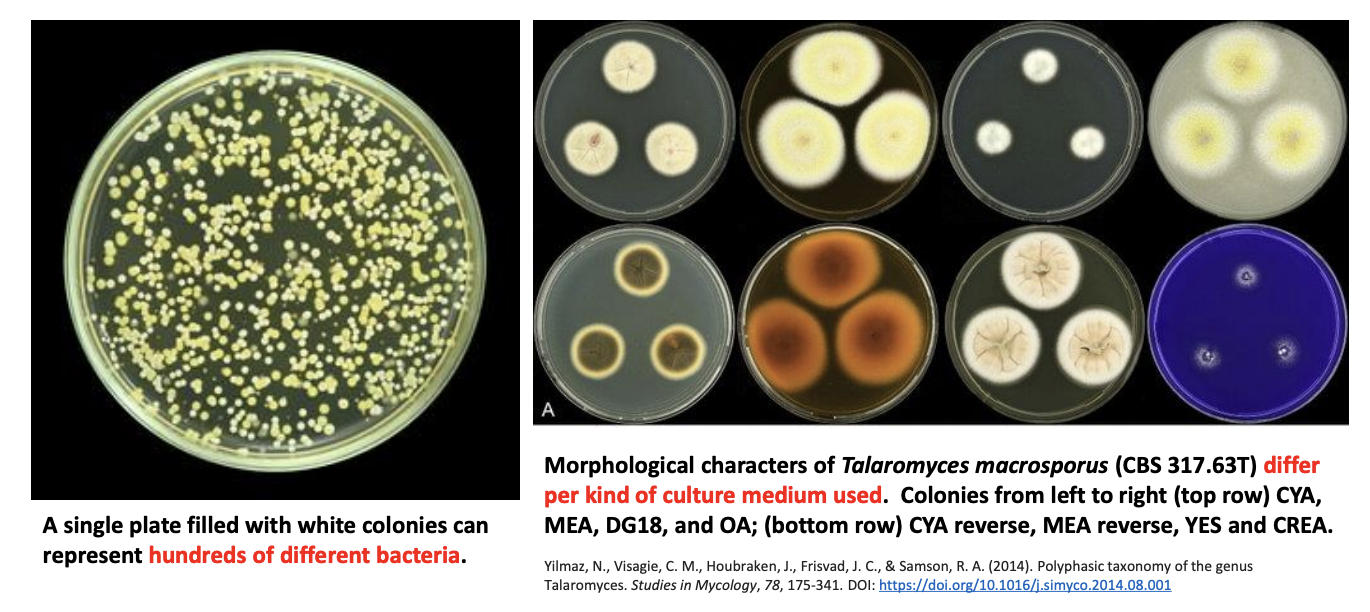
Explain why changing phenotype is a challenge in estimating microbial diversity
Microorganisms are phenotypically indistinguishable or morphologically plastic.
A single plate filled with white colonies can represent hundreds of different bacteria.
Morphological characters of Talaromyces macrosporus differ per culture medium used.
Explain why functional role similarity is a challenge in estimating microbial diversity
Distantly related organisms can fill equivalent functional roles in varied ecosystems.
e.g., Kimchi from different areas have similar but not identical microbiomes, which suggests that different (sometimes even phylogenetically unrelated) microorganisms in a community perform the same fermentation role in that assemblage.
Lactococcus
Lactobacillus
Leuconostoc
Weisella
Explain how horizontal gene transfer poses challenge in estimating microbial diversity
Microorganisms can exchange genes via HGT
Comparative genomic analyses of closely related microbes that exhibit different phenotypes revealed distinct genome differences, which may have come from HGT.
HGT
Transformation: uptake of naked DNA from environment (donor cell lyses)
Transduction: via bacteriophages
Conjugation: via direct contact
Explain how habitat specialization contributes to challenges in estimating microbial diversity
The distribution of some species is restricted, making them rarer to sample.
More heterogeneous or disturbed environments tend to be occupied by generalists
Filter: Interaction between taxa
Specialists tend to develop more in homogeneous or constant environment and determined by natural selection
Abiotic factors: soil composition, pH, moisture
Biotic factors: presence of a host
T/F: Specialist species require more specific sampling techniques. If you can’t sample them, they will be very hard to classify.
TRUE
T/F: For microorganism to be classified, they have to be culturable.
TRUE
T/F: The more culturable microorganisms we have, the more we can sequence them, the more we can update databases.
TRUE
Explain how non-culturability is an issue in estimating microbial diversity.
High proportions of microorganisms across most biomes remain uncultured.
High proportions of microorganisms remain unculturable and can only be detected through high-throughput sequencing methods because we cannot perfectly replicate environmental conditions.
Taxonomic assignments at 97% sequence similarities, we can only accurately identify >10% of most microorganisms due to limitations of sequenced barcodes in databases.
T/F: The classification of what belongs under “microbiology” is always in flux.
TRUE
T/F: Techniques in systematic differentiation, classification, and identification of prokaryotes and eukaryotes are different.
TRUE
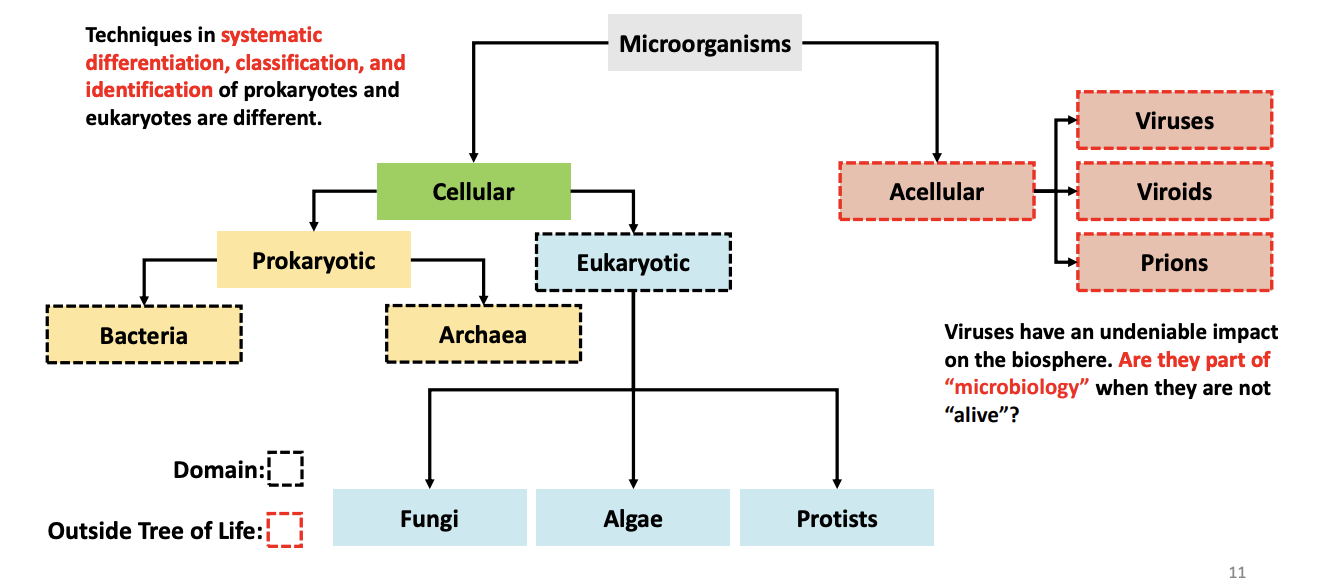
T/F: Bacteria are ubiquitous, mostly free-living, and unicellular prokaryotic microorganisms.
TRUE
Gold standard for prokaryote classification
Gene sequence, not morphology (but it could be something that comes out phenotypically)
Enumerate the 10 key characteristics of bacteria
Cell type
Prokaryotic, peptidoglycan wall, S-layer, glycocalyx
Size
0.5-5 um to 2 cm (Theomargarita magnifica)
Habitat & ecology
Ubiquitous, almost all habitats, extreme environments
Morphology
Unicellular, diverse but with several common forms (cocci, bacilli) and patterns of association (streptococci)
Multicellular forms: aggregates of Myxobacteria, filaments of Actinomycetes, hyphae of Streptomyces, biofilm and microbial mat formations
Metabolism
Great metabolic diversity
Classified based on energy source, carbon source used for growth, electron donors used
Growth & reproduction
Binary fission common, other types are present (budding, fruiting bodies, etc.)
Genetics
Single circular chromosome, plasmid, HGT
Behavior & communication
Flagellar motility in many, pili, bioluminescence, quorom sensing, chemo-, photo-, magneto, and energy taxis
Interactions
Commensals, predators, mutualists, pathogens
Significance
Food and beverage, detergents and enzymes, biotechnology, medicine, bioremediation, pest-control, biogeochemical cycles, etc.
T/F: At species level, 97% genetic similarity is required. Meanwhile, at strain level, 99% genetic similarity is needed.
TRUE
Explain how domain bacteria has constantly changing phyla
There is much consideration to the definition and number of bacterial phyla
Much debate to the definition of phylum in domain bacteria
Common definition: monophyletic lineages sharing ~75% or less of 16s rRNA genes with other phyla
There may be up 1300 bacteria phyla existing
Almost 72% of all bacterial phyla remain “candidates,” i.e., they have no culturable representatives.
41 phyla accepted by List of Prokaryotic names with Standing in Nomenclature (LSPN)
89 phyla recognized in SILVA database
30 phyla have culturable representatives
90% of all culture bacteria belong to either
Bacteroidetes, Proteobacteria, Actinobacteria, Tenericutes, Firmicutes
Almost _ of all bacterial phyla remain “candidates,” meaning they have no culturable representatives
72%
Basis for phylum classification in microbiology
Genetics
The largest and most metabolically diverse bacterial phylum
With most cultured members in record
More than 1/3 of characterized species are found in this group
Constitutes bacteria of medical, industrial, agricultural significance
All gram-negative, with very few metabolism NOT occurring this group
6 classes: Alpha, Beta, Gamma, Delta, Epsilon, Zeta
PROTEOBACTERIA
Second largest class of bacteria
>1000 described species with extensive functional diversity
Most species are obligate/facultative aerobes and many are oligotrophic (can grow in environments w very low nutrient concentrations)
10 well characterized orders
Rhizobiales
Rhizobium: root commensal
Rickettsiales
Rickettsia lysing human cell
Rhodobacterales
Nitrobacter oxidizes NO2- to NO3-
Rhodospirillales
Caudobacterales
Acetobacter produces acetic acid industrially
Sphingomonadales
degrades aromatic organics, performs degradation
Alphaproteobacteria
Third largest class of bacteria
~500 described species with extensive functional diversity
Broad variety of metabolic activities from obligate parasite to living in oligotrophic ground water
6 major orders
Burkholderiales
Hydrogenophilales
Methylophilales
Neisseriales
Nitrosomonadales
Rhodocyclales
Zoogloea ramigera degrades carbon, causes flocculation of wastewater treatment
Neisseria has pathogenic species (Gonorrhea, Meningitis)
Nitrospira are ammonia-oxidizing bacteria
Betaproteobacteria
Alphaproteobacteria that degrades aromatic organics, performs degradation
Sphingomonas
Alphaproteo oxidizes NO2 to NO3
Nitrobacter
Alphaproteo produces acetic acid industrially
Acetobacter
Alphaproteo lysing human cell
Rickettsia
Betaproteo that degrades carbon, causes flocculation of wastewater treatments
Zoogloea ramigera
Betaproteo with pathogenic species causing Gonorrhea, Meningitis
Neisseria
Betaproteo that are important ammonia-oxidizing bacteria
Nitrospira
Largest, most diverse, most culturable bacterial class, most studied, most media formulated with it in mind
Nearly half of characterized species in phylum (>1500)
Rapidly grow in lab media, can be isolated in wide diversity of habitats
Many are pathogenic to animals, humans, plants
15 major orders but most well-characterized are
Enterobacteriales
Shigella dysenteriae invades epithelial cells and can cause bacillary dysentery
Pseudomonadales
Pseudomonas aeruginosa can readily colonize surfaces and cause hospital nosocomial infections
Vibrionales
Vibrio parahaemolyticus inhabits marine habitats and causes gastroenteritis
Gammaproteobacteria
Gammaproteo inhabiting marine habitats, causes gastroenteritis
Vibrio parahaemolyticus
Gammaproteo readily colonizing surfaces, causes hospital nosocomial infections
Pseudomonas aeruginosa
Gammaproteo invading epithelial cells, causes bacillary dysentery
Shigella dysenteriae
3 classes under proteobacteria containing least number of species and functional diversity
Delta-, Epsilon-, Zetaproteobacteria
Proteobacteria
Oxidizes H2S produced by sulfate- and sulfur-reducers
Epsilonproteobacteria
Proteobacteria that contains only 1 characterized species, iron oxidizer Mariprofundus ferrooxydans, but 28 potential others
Zetaproteobacteria
Proteobacteria mostly sulfate- and sulfur-reducing, dissimilative iron-reducing, bacterial predators
Deltaproteobacteria
Causative agents of gastroenteritis, gastritis
Helicobacter, Campylobacter
Common sulfate-reducing microbes
Desulfovibrio, Deltaproteobacteria
Produces mineralized twisted stalks of iron due to Fe oxidation
Mariprofundus ferrooxydans
_ combined contain nearly 50% of all characterized species
Firmicutes, Tenericutes, Actinobacteria
Actinobacteria include _, which is a huge group of primarily filamentous soil bacteria with high C+G concentrations in DNA
Actinomycetes
High C+G conc > higher melting point in DNA bc C+G has 3 H bonds as opposed to A+T (2 H bonds) > important when classifying
_ lacks cell wall, e.g., _ that cannot synthesize peptidoglycan
Tenericutes
Mycoplasma (lack of cell wall contributes to their being highly infectious)
_ includes endospore-forming bacteria, lactic-acid producing bacteria, and other groups, with low C+G concentrations in their DNA
Firmicutes
Bacillales, Clostridiales
Lactobacillales
3 classes under Firmicutes
Lactobacillales, Bacillales, Clostridiales
2 classes under Tenericutes
Mycoplasmatales, Entomoplasmatales
3 classes under Actinobacteria
Actinomycetales, Bifidobacteriales, Coriobacteriales
Firmicutes important in plant food fermentation
Leuconostoc mesenteroides
Firmicutes used in cheese fermentations to produce flavor
Streptococcus thermophilus
Firmicutes extensively studied as cell factory for protein products
Lactococcus lactis
Firmicutes used commonly in making yoghurts
Lactobacillus delbrueckii subsp. Bulgaricus
Class under Firmicutes containing industrially significant lactic acid bacteria
Lactobacillales
Genera of bacteria composed of aerotolerant obligate fermenters important to various industries
Lactic Acid Bacteria (LAB) group
T/F: All lactic acid bacteria produce lactic acid as major or sole fermentation product and, thus, can be divided into:
Homofermentive (only produce lactic acid)
Heterofermentive (produce lactic acid, as well as smth else, e.g., ethanol, propionic acid)
TRUE
T/F: Most LABs can obtain energy only from sugar fermentation which expands the habitats where they are isolated.
FALSE
Most LABs can obtain energy only from sugar fermentation which restricts habitats where they are isolated.
T/F: Many members of Clostridiales and Bacillales do not form spores.
TRUE
3 examples of nonspore-forming genera
Listeria
Staphylococcus
Sarcinia
Facultative aerobe that can grow fermentatively
Can tolerate drying and high salt concentrations
Common commensals in animals, humans but can cause serious illness (MRSA)
Staphylococcus
Found widely in soil, opportunistic pathogen
Common cause of foodborne illness
Transmitted through ready-to-eat food, e.g., cheese, sausages, which are cold-tolerant
Listeria
Obligate anaerobes that divide in 3 perpendicular planes to create packets for 8 or more cells
Can be isolated from soil, mud, feces, stomach contents due to its extreme acid tolerance
Sarcina, e.g., Sarcinia ventriculi in mucosa of gastric ulcer patient
T/F: All endospore-forming bacteria are Gram-negative species of Bacillales or Clostridiales
FALSE
Gram-positive species of Bacillales, Clostridiales (BLUE to PURPLE)
3 spore-forming genera
Bacillus, Paenibacillus
Clostridium
Sporosarcina
Spore-forming genera that produces extracellular enzymes that break biopolymers, produces antibiotics during stationary phase, produces toxic insecticidal proteins
Bacillus, Paenibacillus
Spore-forming genera whose cells are cocci not rods; catabolizes urea into ammonia, raising soil pH; uniquely cultured until pH 10
Sporosarcina
Spore-forming genera that lives in anoxic pockets of soil, lacks respiratory chain, uses substrate-level phosphorylation
Produces butyric acid, acetone, butanol from sugars and polymers, e.g., cellulose
Ferment amino acids
Produce toxins deadly to humans, e.g., C. tetani, botulinum, perfringens
Clostridium
_ could cause muscular contractions strong enough to break bones (backbreaker disease)
Tetanus
_ produces insecticidal parasporal protein (X)
Bacillus thuringiensis
T/F: Bacillus and Paenibacillus become highly active in producing enzymes BEFORE entering sporulation stage.
TRUE
This increase in enzyme production helps in breaking down organic matter in their environment, which is beneficial for nutrient acquisition and survival.
T/F: Stressful conditions induce Bacillus & Paenibacillus to enter a specific growth phase where antibiotic production is triggered.
TRUE
Antibiotic production is a defense mechanism to reduce competition, particularly during periods when resources are very limited.
Normally, Agrobacterium AT has pathogenic DNA that causes a type of disease in plant called _
Crown gall disease
Explain how we make BT corn, rice, etc.
Isolate gene encoding insecticidal parasporal protein from BT
Insert BT gene into plasmid
Transform plasmid into Agrobacterium tumefaciens (AT)
Infect plant cells with transformed Agrobacterium AT cells
Agrobacterium tumefaciens causes a type of plant disease known as crown gall, which results from the insertion of its own DNA into the plant's genome. Scientists exploit this mechanism by replacing the pathogenic DNA with the BT gene.
C. tetani vs. C. botulinum
Tetani: continuous muscle contractions, spasms
Botulinum: continuous muscle relaxation, paralysis
Dormant, tough, asexual, nonreproductive structures that have high and wide ranging tolerance to harsh climactic conditions. Its production require genes not acquired via HGT.
Endospore
T/F: Production of endospores require genes acquired via HGT
FALSE
Production of endospores require genes NOT acquired via HGT
Single class under Tenericutes lacking bacterial cell wall
Mollicutes
Latin for soft
Lacks cell walls due to adaptation of living as symbionts, intracellular pathogens
Some of the smallest microbes
Mollicutes
Mollicutes genera
Most well-characterized genus
Sterols in cell membrane for protection against osmotic lysis
Lipoglycans for added protection, adherence to surfaces
Have varied growth morphology in media
Mycoplasma
_ (TEM) has varied growth morphology: coccoid, hyphae-like elements
Mycoplasma mycoides
_ detected in hemolymph of Drosophila pseudoobscura, causing it to bear only female progeny
Spiroplasma sp.
Mollicutes genera
Helical, spiral-shaped
Lacks flagella but have rotary, screw motion
Commonly found in gut of insects, internal tissues of plant
Spiroplasma
_ have been detected in plants, suggesting that a large group of plant-associated Mycoplasma may exist
Mycoplasma-like organisms
Phylum rod-shaped to filamentous, primarily aerobic, common inhabitants of soil and plant materials
Many members are commensals, except Mycobacterium, which has pathogenic members
Some members are economically significant as producers of antibiotics or fermented dairy products
Contains 9 orders but most species belong to Actinomycetales (Gram-positive, anaerobic, have “mycelia”)
Actinobacteria
Actinobacteria species
Have unusual filamentous branching growth pattern
Actinomyces israelii
Actinobacteria species
Have critical role in acne formation
Propionibacterium acnes
Actinobacteria species
Isolated from AIDS patient
Mycobacterium avium
Colonies of _ and other soil bacteria in casein-starch agar plate
Streptomyces
Have an unusual method of cell division
Gram-positive, aerobic, nonmotile, rod-shaped organisms; irregularly shaped, club-shaped, v-shaped cell arrangement during growth
V-shaped cells arise from snapping division, where outer layer of cell wall of diving cell lyses while inner layer remains fused
Coryneform bacteria
Main genera of coryneform bacteria
Corynebacterium, composed of animal & plant pathogens, saprophytes
Arthrobacter, nutritionally versatile & stress-resistant microorganisms