control of gene expression bio 329\
1/95
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
96 Terms
All cells of a multicellular organism contain the same
Genome
What is a genome
An entire set of DNA of an organism
All cells posses the same genome but express only the
RNA and proteins specific for that cell type (differentiation)
Differentitated cells contain all the
Genetic information of the organism
Difference cell types produce different set of
Proteins
Many proteins are common to all cells of a
Multicellular organism
Housekeeping proteins used for
Common functions of cells
Each cell type also produces
Specialized proteins that are responsible for the cell’s distinctive properties
Eukaryotic cells can control which proteins they contain by
regulating various steps along the pathway to gene expression.
most genes the main site of control is
step 1: transcription of a DNA sequence into RNA.
Many proteins require what to become fully activated
Post translational modifications
Transcription regulator binds to
Regulatory DNDA sequences
How is transcription regulated
Through activators, repressors, and inducers
Repressors turn genes
Off
Activators turn genes
On
Repressors and activators
are proteins that bind DNA and regulate expression
Inducers and co-Repressors are small molecules that may participate in
Gene regulation by altering the function of repressors and activators
Prokaryotic genes are often organized into
Operó a
Operons are
Cluster of genes that are transcribed as a single unit
Operon allow regulation of
Expression of The entire cluster through a single transcription switch
Switches allow cells to respond to changes in their
Transcription environment
A cluster of bacterial genes can be transcribed from a single
Promoter
Repressor proteins regulate
Trp operon gene expression
If the concentration of tryptophan inside a bacterium is low
RNA polymerase binds to the promoter and transcribes the 5 genes of the tryptophan operon
However, if the concentration of tryptophan is high
the repressor protein becomes active and binds to the operator, where it blocks the binding of RNA polymerase to the promoter.
When the concentration of intracellular tryptophan drops, the repressor falls off the
DNA, allowing the polymerase to again transcribe the operon. The promoter contains two key blocks of DNA sequence information, the –35 and –10 regions,
The -10 and -35 regions are recognized by
RNA polymerase
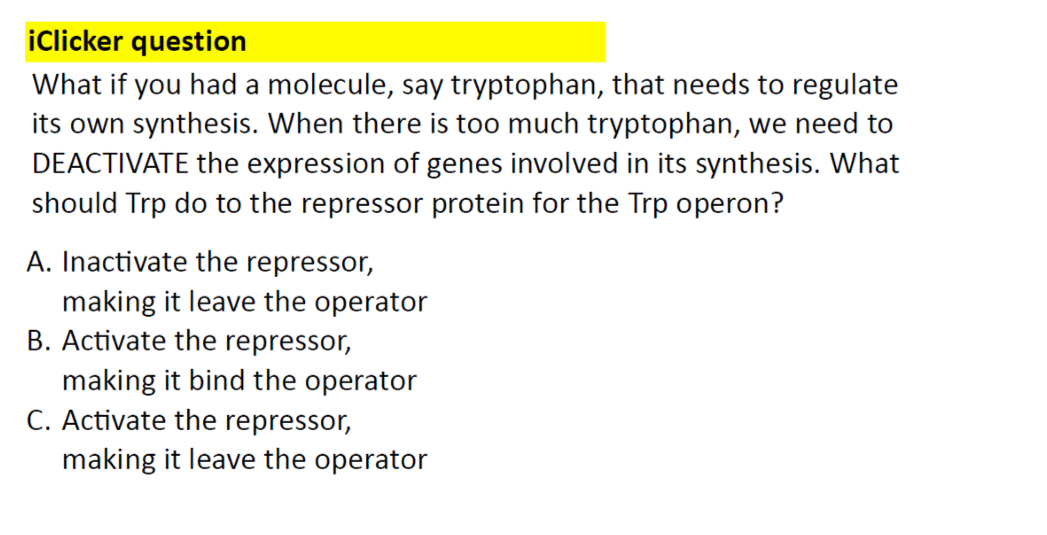
Activate the repressors making it bind to the operator
The lac operon is controlled by two transcription regulators:
The lac repressor and CAP
When lactose is absent, the Lac repressor binds to the
LAC operator and shuts off expression of the operon
allolactose binds to the Lac repressor, causing it to undergo a conformational change that releases
its grip on the operator DNA
When glucose is absent cyclic AMP and CAP does what?
Cyclic AMP is produced by the cell and CAP binds to the DNA
For the operon to be transcribed, glucose must be
Absent and lactose must be present
LacZ, the first gene of the operon, encodes the enzyme
β-galactosidase, which breaks down lactose into galactose and glucose

Inactivate the repressors, making it leave the operator
Transcription is controlled by
Proteins binding regulatory DNA sequences
Regulatory proteins are
Specific transcription factors
Regulatory proteins bind DNA and either stimulate or inhibit the
RNA polymerase
Regulatory proteins binding are achieved by interactions between
Protein and DNA
In enkaryotes, gene activation can occur at a
Distance
An activator protein bound to a distant enhancer attracts
RNA polymerase and the general transcription factors to the promoter.
Looping of the intervening DNA permits contact between the
activator and the transcription initiation complex bound to the promoter.
The arrangement of chromosomes into looped domains keep Enhancers
In check
Eukaryotic genes are controlled by
Combinations of transcription regulators
DNA sequences may play a structural role in allowing the regulatory DNA sequences to find their proper positions, but they are not recognized by any
Transcription regulators
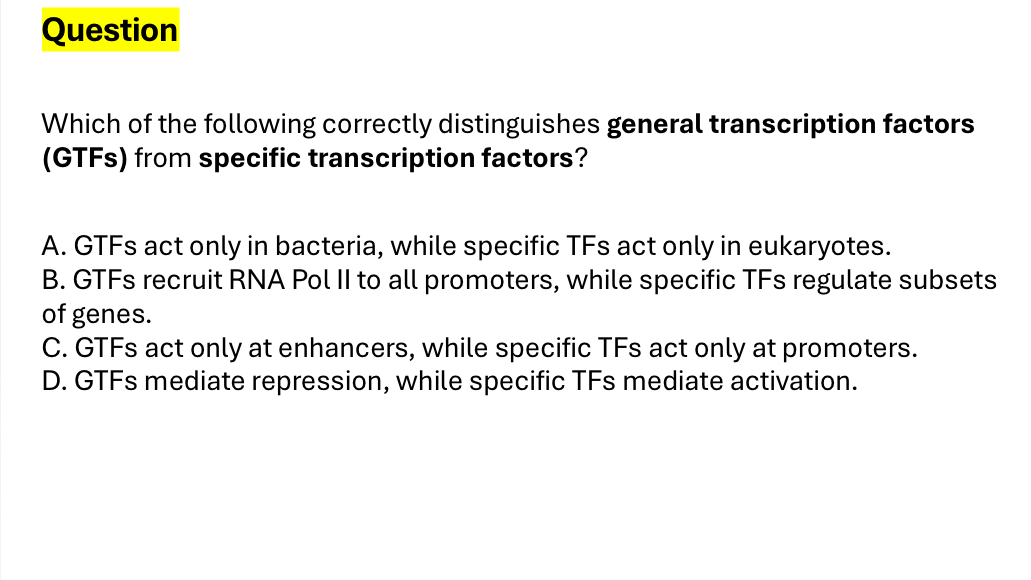
GTF recruits RNA pol II to all promoters, while specific TF regulate subsets of genes
Combinations of a few transcription regulators can generate
Many cell types during development
The expression of different genes can be coordinated by a single
Protein
Interphase chromosomes contain both highly condensed and more extended forms of
Chromatin called euchromatin and heterochromatin
Euchromatin
Extended, more loosely package, accessible for gene expression
Heterochromatin
Tightly package, not expressed
Facultative heterochromatin are
Regions of tightly packaged DNA that can be modified to switch to euchromatin
Constitutive heterochromatin are
Always tightly packed, not expressed, often in structural regions of the chromosomes
Eukaryotic transcriptional activators can recruit chromatin-modifying proteins to help
initiate gene transcription
What are the mechanism of gene regulation by TF
Coactivators and corepressors
TFs can recruit
Histone acetlytransferanse (HATs)
HATs add acetylene groups to
Lysines in histone tails
Histones acetylation reduces the affinity of
Histones to DNA
Gene activation allows Transcriptional machinery to access
DNA
In general silencing TFs recruit
Histone deacetylase (HDACs)
HDACs remove acetylene groups from
Histones
Gene silencing silences
Gene expression
In DNA methylation the addition of a methyl group is added on the
5’-C of cytosine base
DNA methylation is carried out by
DNA methyltransferases
Up to 10% of the cytosines in vertebrates are
Methylated
DNA methylation leads to
Gene silencing
Methylated promoters are less likely to bound by
PIC proteins
DNA methylation patterns can be faithfully inherited when a cell
Divides
An enzyme called a maintenance methyltransferase guarantees that once a pattern of DNA methylation has been established, it is inherited by
newly synthesized DNA
Immediately after DNA replication, each daughter double helix will contain
one methylated DNA strand—inherited from the parent double helix—and one unmethylated, newly synthesized strand
The maintenance methyltransferase interacts with these hybrid double helices and methylates only those
CG sequences that are base-paired with a CG sequence that is already methylated
Cytosine ca be methylated in
DNA
mRNAs Contain Sequences That Control Their
Translation
Regulatory RNAs Control the Expression of Thousands of
Genes
MicroRNAs Direct the Destruction of
Target mRNAs
Small interfering RNAs protect cells from
Infections
Bacteria use small non coding RNAs to protect themselves form
Viruses
Thousands of Long Noncoding RNAs May Also Regulate
Mammalian Gene Activity
Eukaryotic genes contain many
Exons
mRNA can retain all or splice some of __ out
Exons
In alternative splicing the same gene can produce different
mRNAs
Splicing pathways may depend on the
Stage of development, cell type, and tissue type
bacterial gene’s expression can be controlled by
Regulating translation of its mRNA
Sequence-specific RNA-binding proteins can repress the translation of specific mRNAs by
keeping the ribosome from binding to the ribosome-binding sequence in the mRNA
If a ribosomal protein is accidentally produced in excess over other ribosomal components, the free protein will
inhibit translation of its own mRNA, thereby blocking its own synthesis.
As new ribosomes are assembled, the levels of the free protein
Decrease, allowing the mRNA to again be translated and the ribosomal protein to be produced
An miRNA targets a complementary mRNA molecule for
Destruction
Each precursor miRNA transcript is processed to form a double-stranded intermediate, which is further processed to form a
mature, single-stranded miRNA
miRNA assembles with a set of proteins into a complex called
RISC, which then searches for mRNAs that have a nucleotide sequence complementary to its bound miRNA
Depending on how extensive the region of complementarity is, the target mRNA is either rapidly
degraded by a nuclease within the RISC or transferred to an area of the cytoplasm where other nucleases eventually destroy it
Preston’s may be selectively targeted for degradation when there are
Misfolded or damaged proteins and regulatory proteins that need to be removed
Proteins destined for degradation are labeled with
Ubiquitin; a small protein
Ubiquitinated proteins bind to the cap of
Proteasome, a huge proteolytic complex
Proteins are sucked in to the
Proteasome and is destroyed
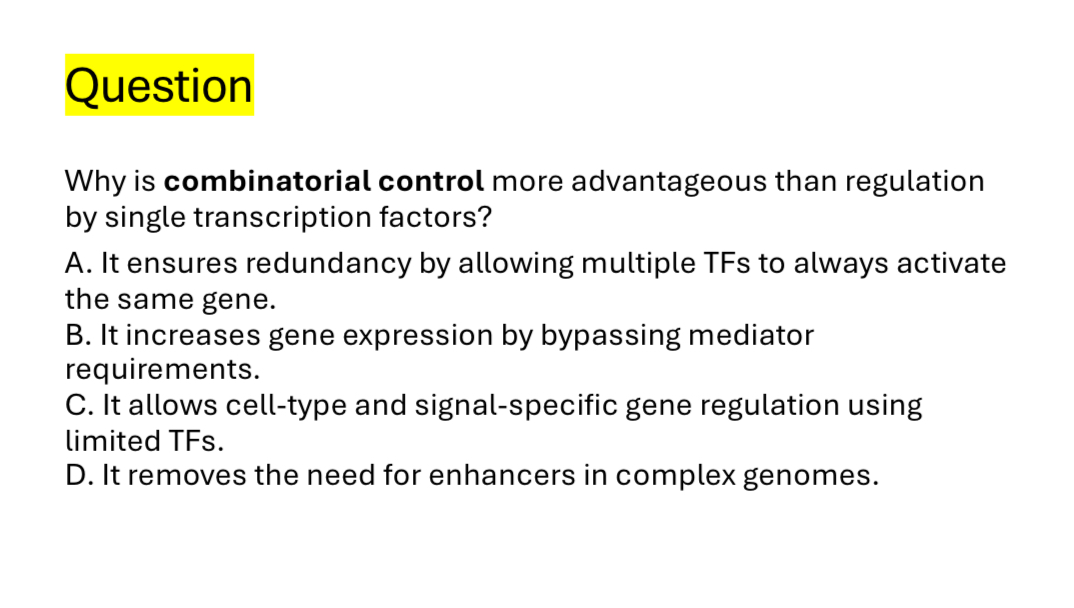
It allows cell types and signal specific gene regulation using limited TFs
What are the importance of regulation of gene expression
By altering gene expression, organisms can adapt to environmental challenges.
Transcription control can result in tissue specific gene expression.
Gene regulation is influenced by hormones, heavy metals and chemicals.
Dysregulation of gene regulation can lead to disease.