BIOL10008: Transcription, Translation, Mutations
1/32
Earn XP
Description and Tags
Concepts 13 and 14
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
33 Terms
Transcription
DNA sequence info is copied into a complementary RNA sequence by RNA polymerase
Occurs in nucleus
Initiation, elongation, termination
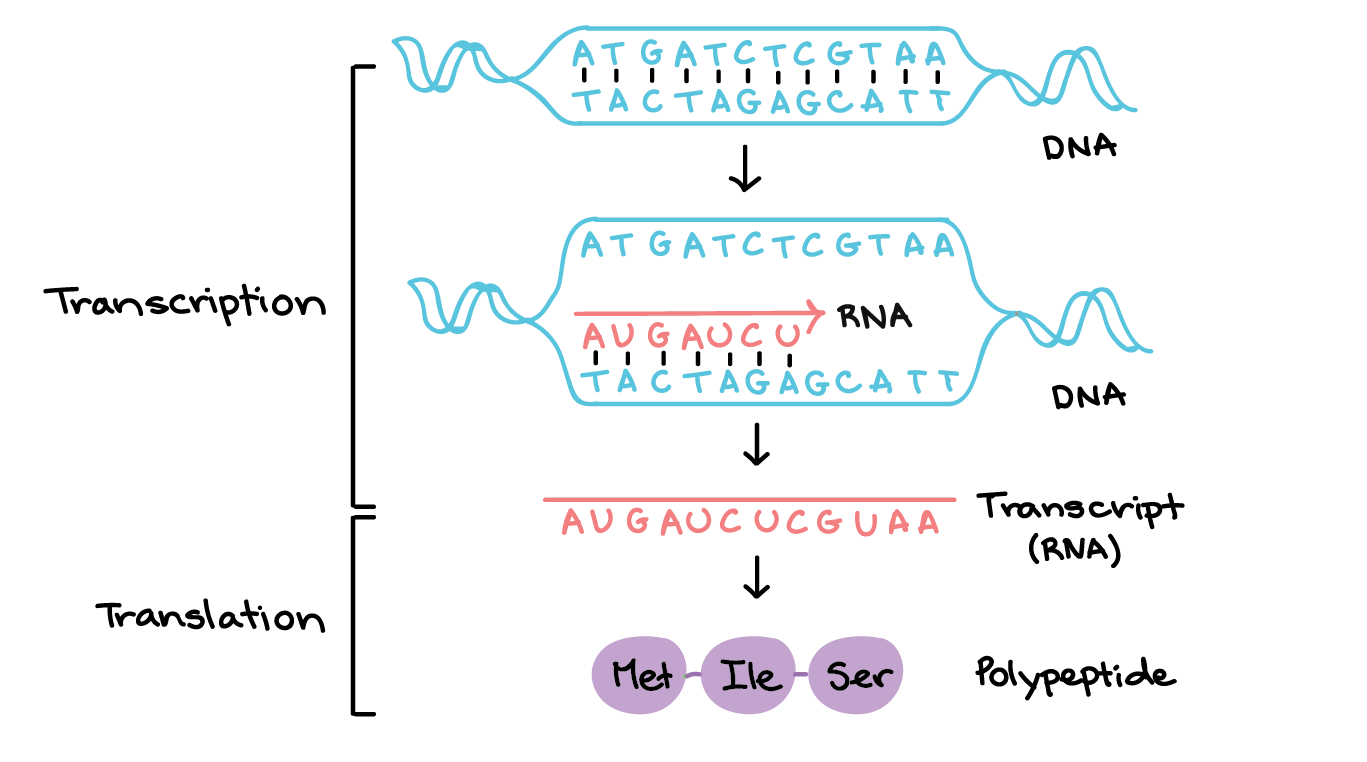
What is the coding strand and template strand?
Template strand (non-coding strand) - complementary to the mRNA strand, runs from 3’ to 5’
Coding strand (called the complementary strand) - complementary to template strand, identical to mRNA (T instead of U), 5’ to 3’
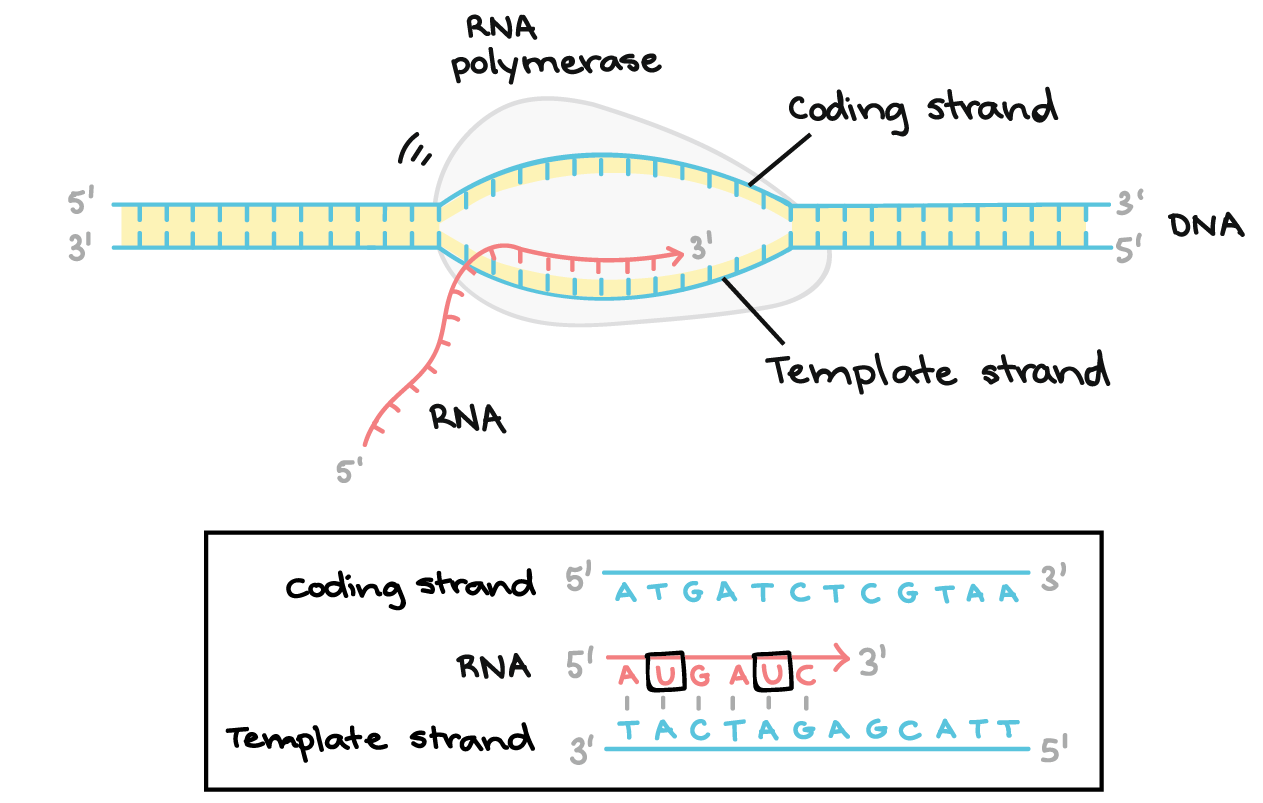
Describe the first step in transcription
Initiation
RNA polymerase binds to the promoter and unwinds the double-stranded DNA
The promoter indicates where transcription begins (initiation site) and which strand to transcribe.
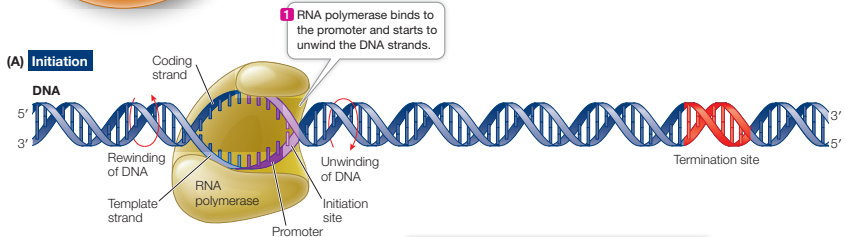
How is RNA polymerase assisted with binding to the promoter?
Promoter has directionality, so RNA polymerase needs to be orientated to bind
groups of nucleotides upstream (towards 3’ of template strand) of the initiation site help it bind
sigma factors in prokaryotes and transcription factors in eukaryotes help direct the enzyme onto the promoter - they recruit RNAP to specific genes
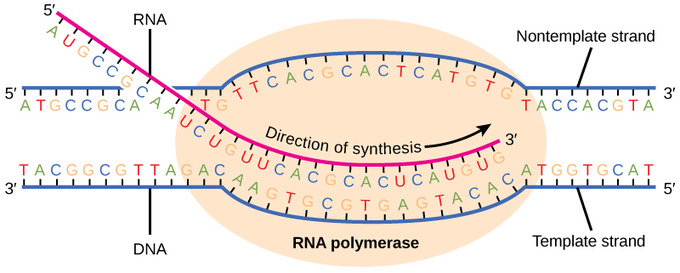
Describe the second step in transcription
Elongation
DNA strand continues to unwind, and RNAP reads the template strand in the 3’ to 5’ direction
As it reads the template strand, RNAP synthesises the RNA molecule by adding complementary nucleotides to a chain that grows from 5’ to 3’
The RNA transcript is antiparallel to the DNA strand
Complementary nucleotides are of the bases adenine, guanine, cytosine and uracil
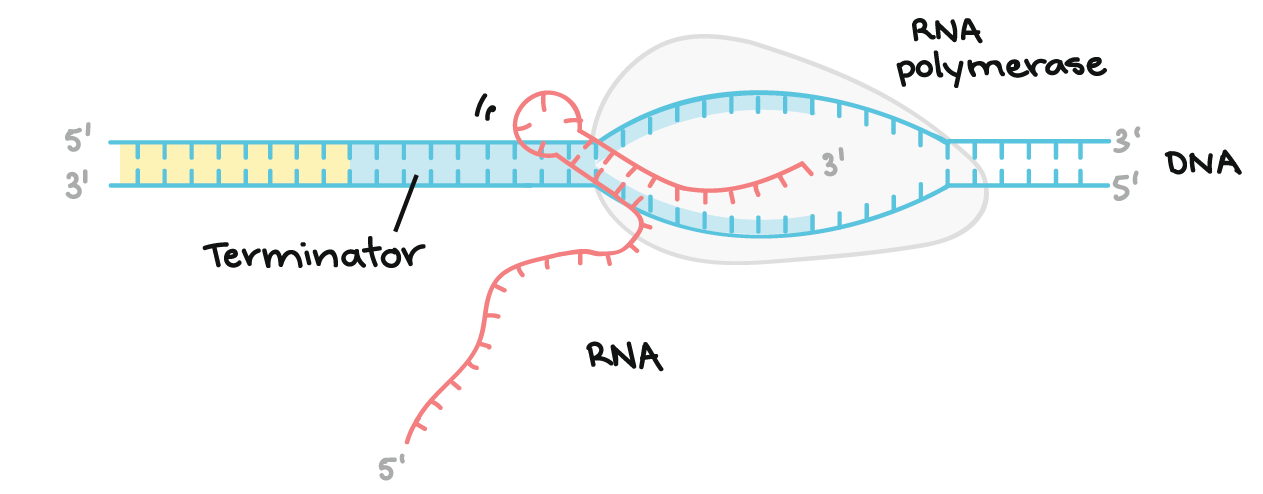
Describe the third step in transcription
Termination
Once RNA polymerase reaches the termination site (particular base sequences) on the DNA, a mechanism occurs that detaches the transcript and DNA from RNAP
RNA transcript is complementary to template strand and identical to coding strand (uracil base instead of thymine)
The mechanism in some genes causes the transcript to fold and form internal hydrogen bonds with itself, forming a hairpin loop. This loop causes the transcript to fall away from the DNA and RNAP. In other cases, a protein binds to specific sequences on the transcript and causes the RNA to detach from the DNA template
Differences between Prokaryotic and Eukaryotic Gene Expression
Transcription and Translation Occurrence:
Prokaryotes - at same time in cytosol; Eukaryotes - transcription in nucleus, translation in cytosol
Precursor mRNA Processing:
Prokaryotes - usually none; Eukaryotes - introns spliced, 5’ end cap and 3’ poly A tail added (these modifications cause differences in gene structure between DNA and mature RNA, regions won’t align)
Why doesn’t RNA processing occur in prokaryotes?
In prokaryotes, the sequence of the mRNA strand is 1:1 complementary with the DNA template strand due to transcription and translation both occurring in the cytosol at the same time
In eukaryotes, the mRNA strand contains introns (non-coding regions) which need to be spliced
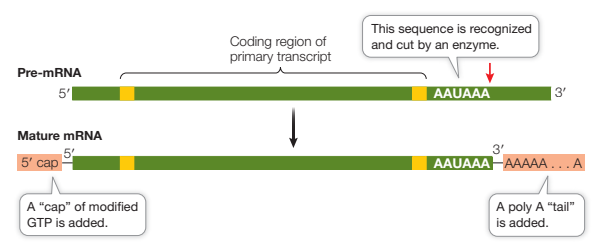
What modifications occur in RNA processing?
In the nucleus:
RNA splicing removes introns and splices exons together through recognition of certain sequences
5’ “cap” of GTP is added which facilitates the binding of mRNA to the ribosome for translation, and protects the mRNA from being digested by ribonucleases
Poly A tail added to 3’ end
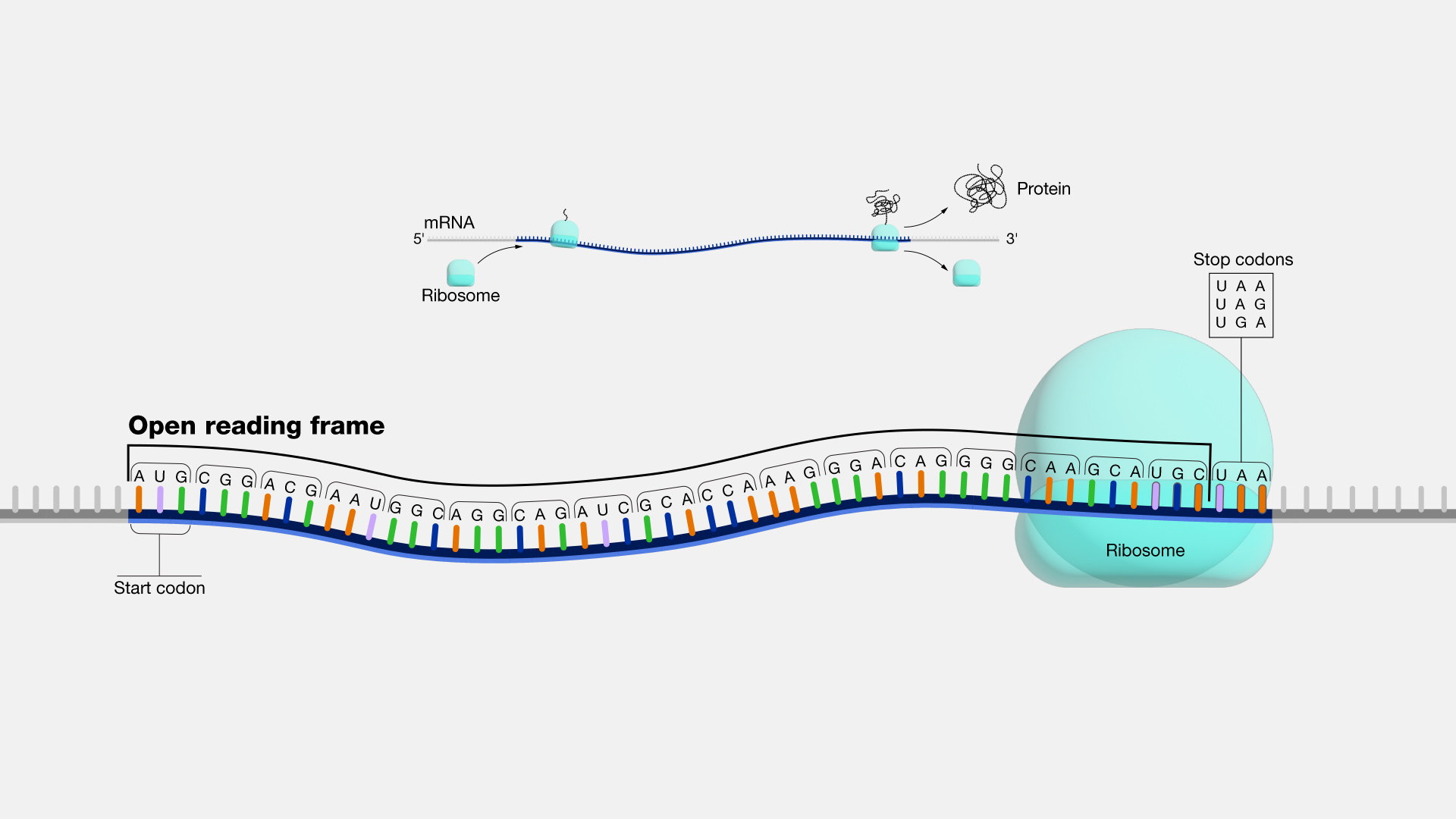
What is an open reading frame? Describe the features
Region of DNA sequence that corresponds to a mRNA transcript and can be translated by a ribosome into a continuous polypeptide chain.
Contains a start codon (AUG), ends with a stop codon (UAA, UAG, UGA)
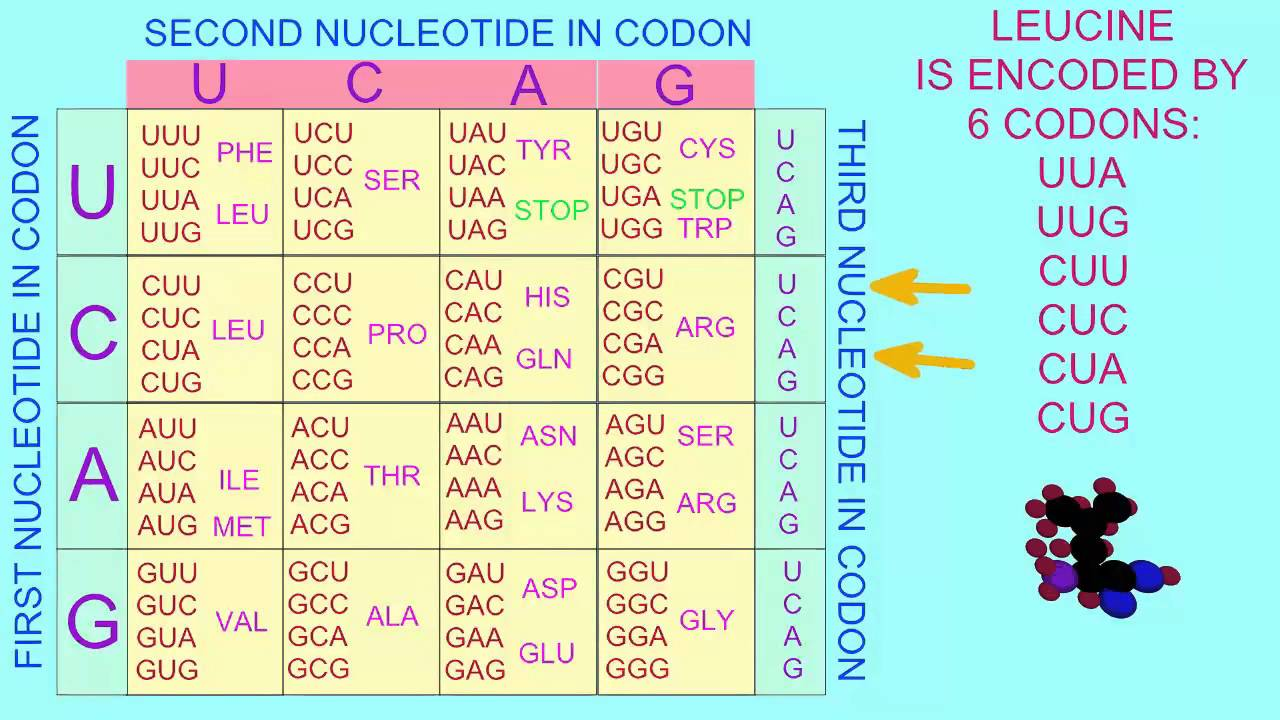
Describe what is meant by degeneracy of the genetic code. What is a possible advantage?
There are 64 possible codons and 20 amino acids that codons can encode for.
The degeneracy/redundancy of the genetic code refers to the fact that multiple codons can encode for most amino acids.
Therefore, silent mutations can occur (substitutions of nucleotide in sequence won’t change the amino acid the codon encodes for)
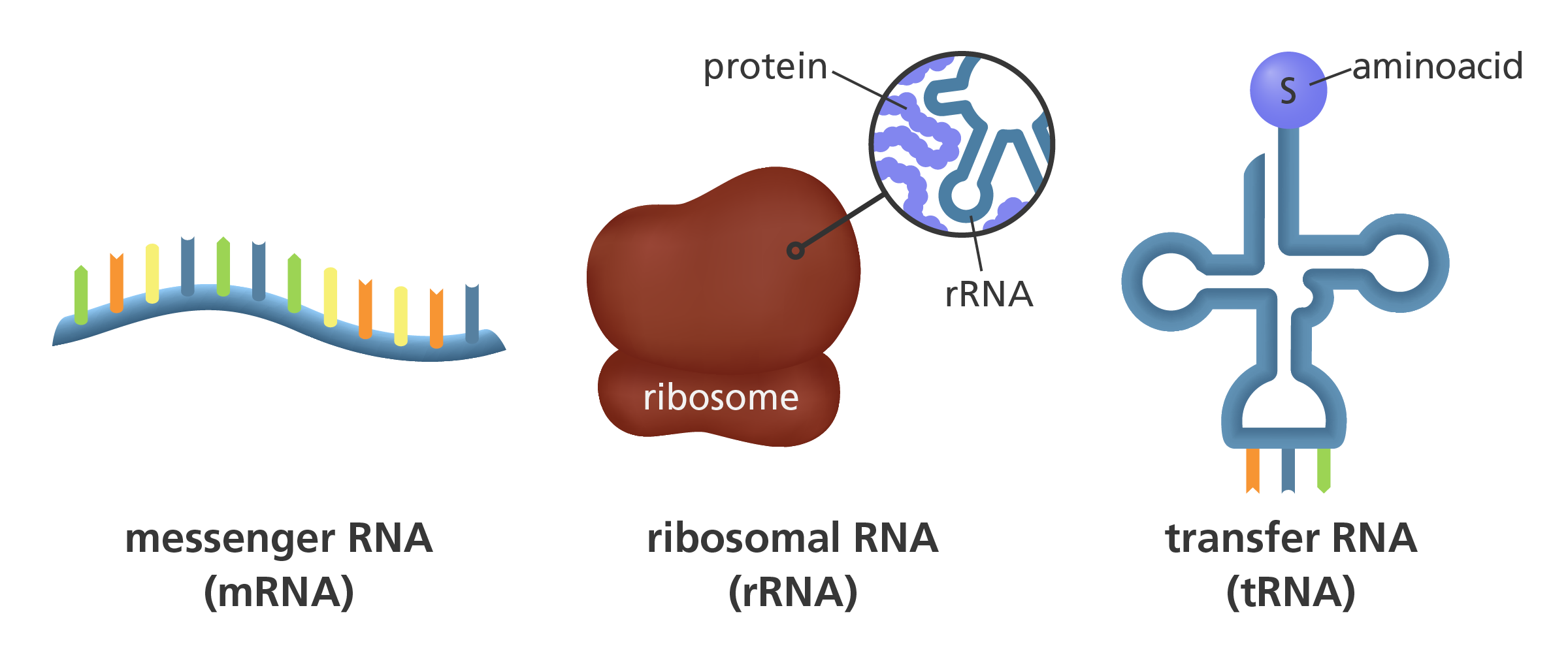
What are the three main types of RNA produced and what are their roles?
Messenger RNA - carries sequence complementary to DNA template strand, travels from nucleus to cytoplasm where protein synthesis occurs, used as a ‘blueprint’
Ribosomal RNA - a part of the ribosome, one of the rRNAs catalyses peptide bond formation between amino acids, to form a polypeptide
Transfer RNA - binds to particular amino acids becoming charged, then binds to mRNA where its anticodon is complementary to the mRNA codon for the specific amino acid being carried, non-covalently interacts with ribosome
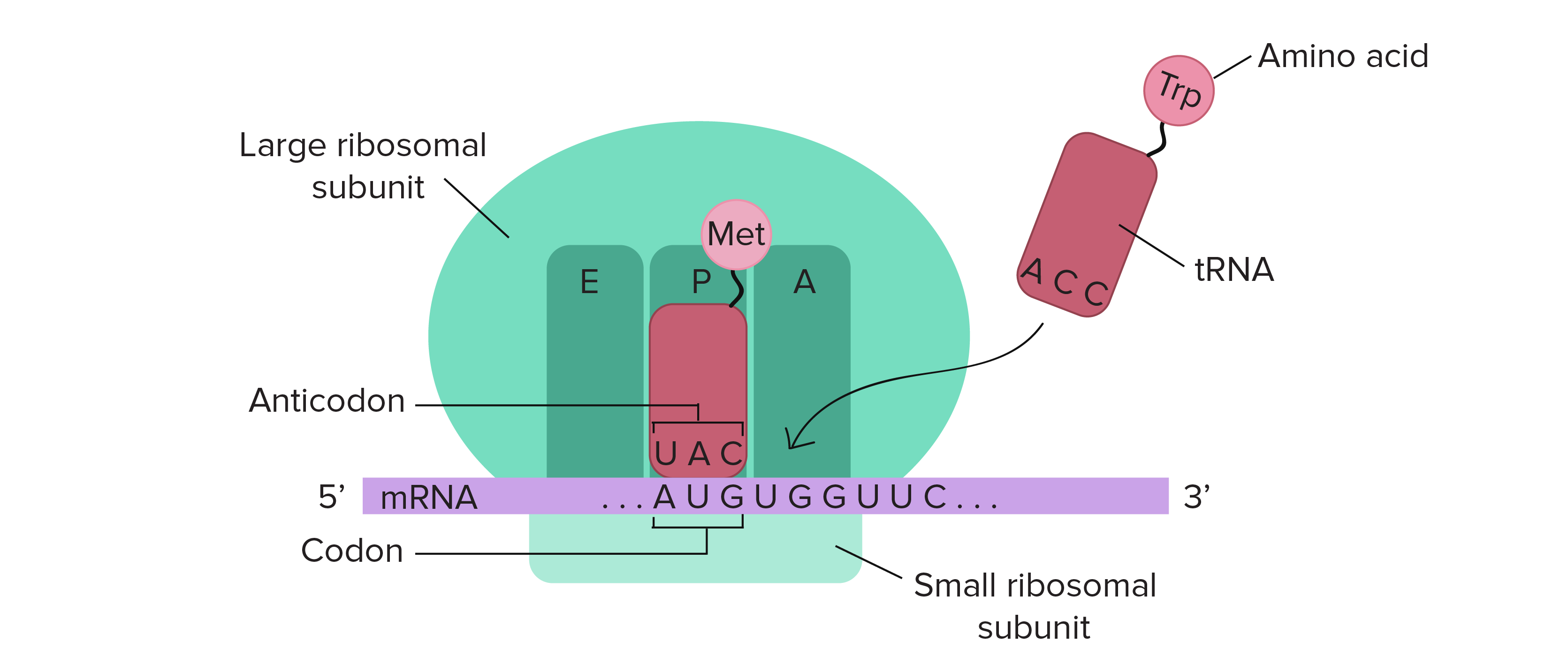
Translation
Facilitated by ribosomes (comprised of large subunit and small subunit)
Synthesis of polypeptide chains from genetic information carried by RNA
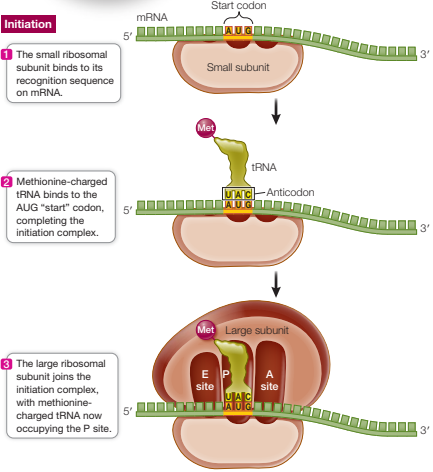
Describe the first phase of translation and how it differs in prokaryotes and eukaryotes.
Initiation
Formation of initiation complex:
Prokaryotes - rRNA of small ribosome subunit binds to Shine-Dalgarno sequence on mRNA
Eukaryotes - small ribosomal subunit binds to the 5′ cap on the mRNA
Small subunit then moves along mRNA until it reaches start codon (AUG)
Met-charged tRNA with anticodon UAC binds to the start codon through complementary base pairing, completing the initiation complex
large ribosomal subunit joins the initiation complex, methionine-charged tRNA now occupying the P site and the A site is aligned with the next codon
Components assembled by initiation factors
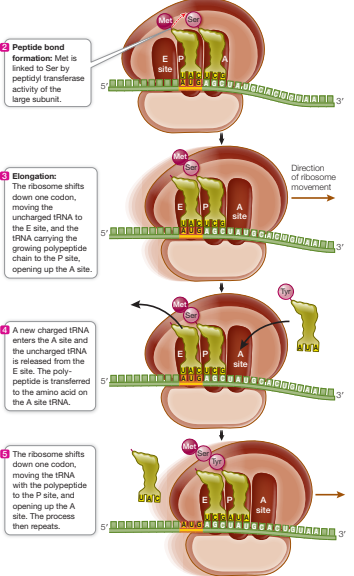
Describe the second phase of translation
Elongation
Charged tRNA enters A site carrying amino acid complementary to codon
Large ribosomal subunit catalyses: the breaking of bond between the tRNA and amino acid in P site, and formation of peptide bond between amino acid from P site and amino acid in A site
Ribosome shifts to next codon, releasing uncharged tRNA from the E site
A site is open and the process repeats
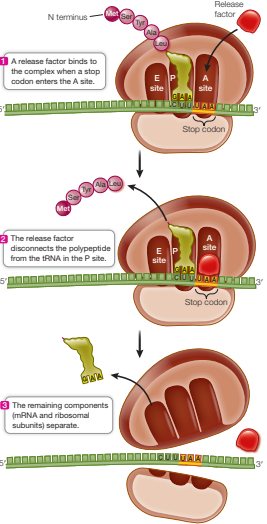
Describe the third phase of translation
Elongation ends when a stop codon (UAA, UAG, or UGA) enters the A site
Stop codon binds with a protein release factor which allows hydrolysis of the bond between the polypeptide chain and the tRNA in the P site
New polypeptide separates from ribosomal subunits
Ribosomal subunits and mRNA separate
Mutations
Change in the nucleotide sequence in DNA that can be passed on from one cell or organism to another
Allows for genetic variation
Define and provide an example of a spontaneous mutation.
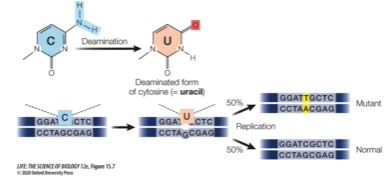
Permanent changes in DNA that occur without any outside influence.
EX: Through deamination (removal of a NH2 amino group) of cytosine base which results in the formation of uracil. If not repaired, in DNA replication DNA polymerase will recognise it as thymine and the mutation will be passed on
Ex: Transposons (DNA element containing sequence) are able to move positions in the genome. Replicative transposition is where a transposon replicates and inserts itself into a new part of the genome - if it is in a part of a gene, it could disrupt protein encoding sequence
What are induced mutations? Name two ways mutations can be induced.
Occur when a mutagen causes a change in DNA leading to mutation
chemicals can alter nucleotide bases (ex: nitrous acid causes deamination)
Some chemicals add groups to the bases (ex: benzopyrene adds chemical group to guanine, making it unavailable for base pairing)
Radiation:
Ionising radiation (X-rays, gamma rays) - produce free radicals which can change bases in DNA to forms that are not recognized by DNA polymerase, also can break sugar-phosphate backbone
Ultraviolet radiation can cause covalent bonds between adjacent thymine bases, blocking DNA replication if unrepaired
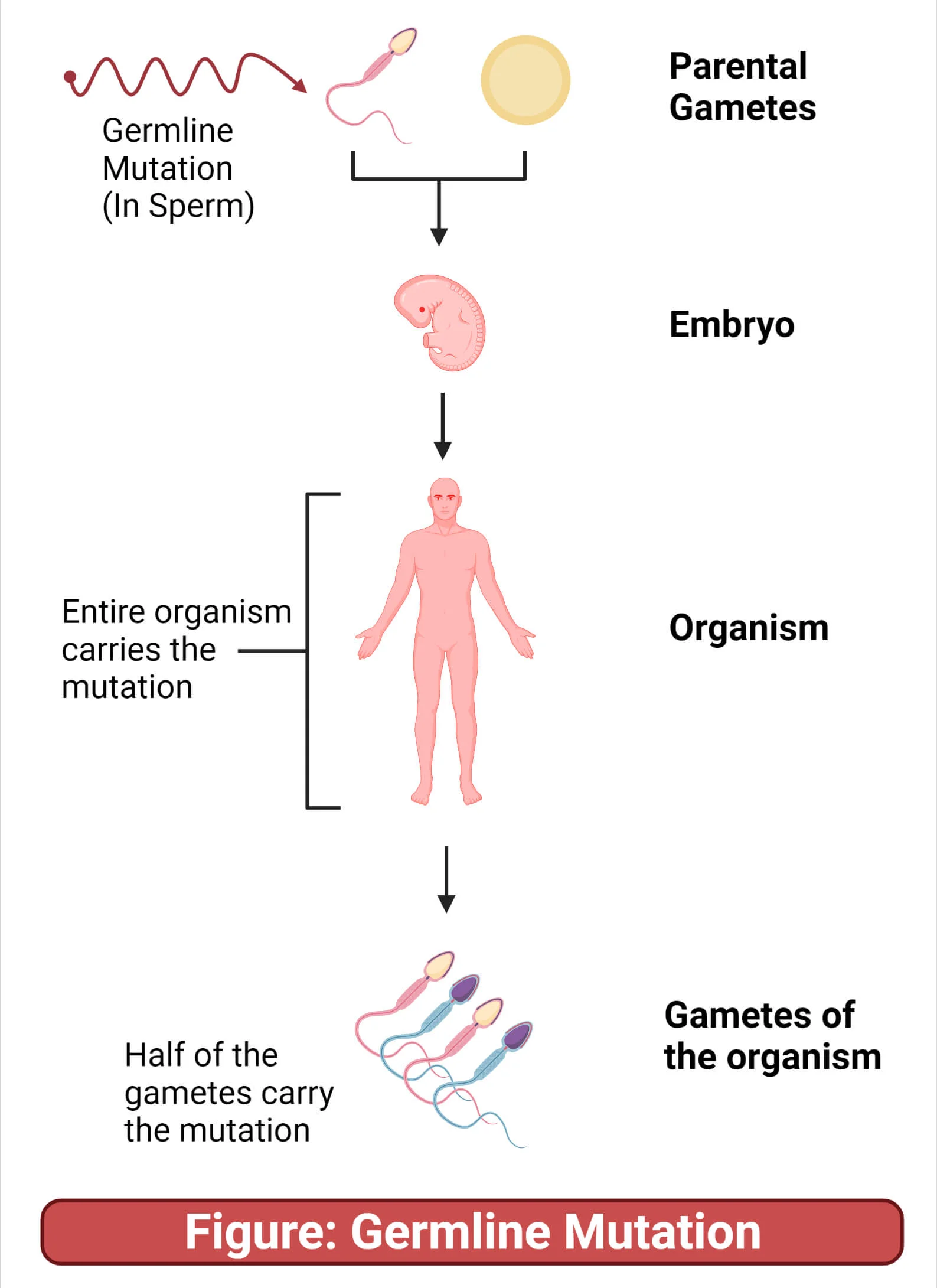
What are germline mutations?
Mutation in germ line cells (cells that give rise to gametes (sperm cells, egg))
Gamete with the mutation will pass it to a new organism in fertilisation
New organism will have mutation in every cell, and will be able to pass it to offspring
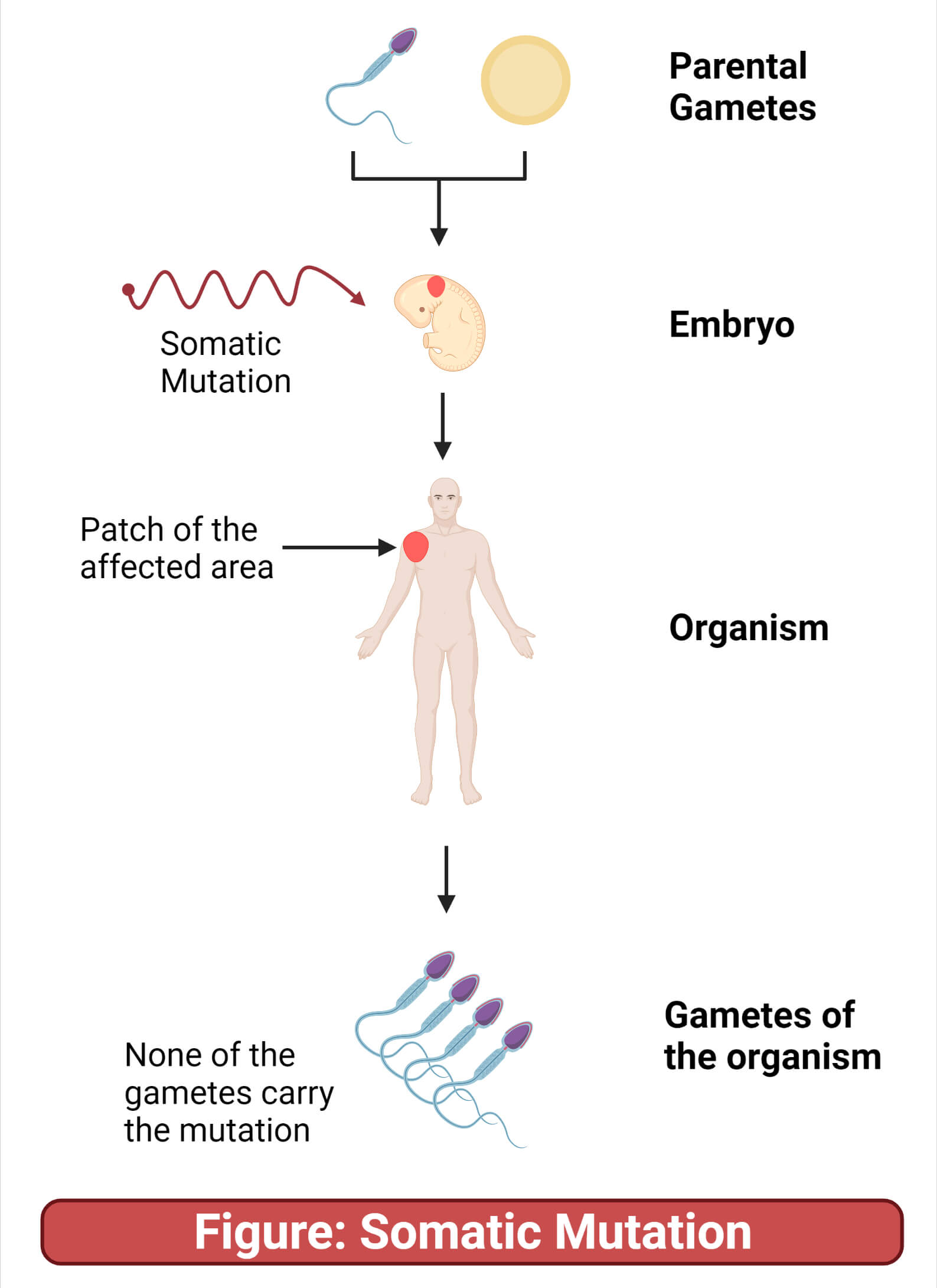
What are somatic mutations?
Occurs in somatic (non-gamete) cells
Mutations passed on to daughter cells during mitosis, and to daughter cells of those cells as well
Not passed on to sexually reproduced offspring
Ex: mutation in a single human skin cell could result in a patch of skin cells that all have the same mutation
List conditions that can induce DNA damage.
Heat
Radiation
Oxidation by free radicals produced during cellular respiration
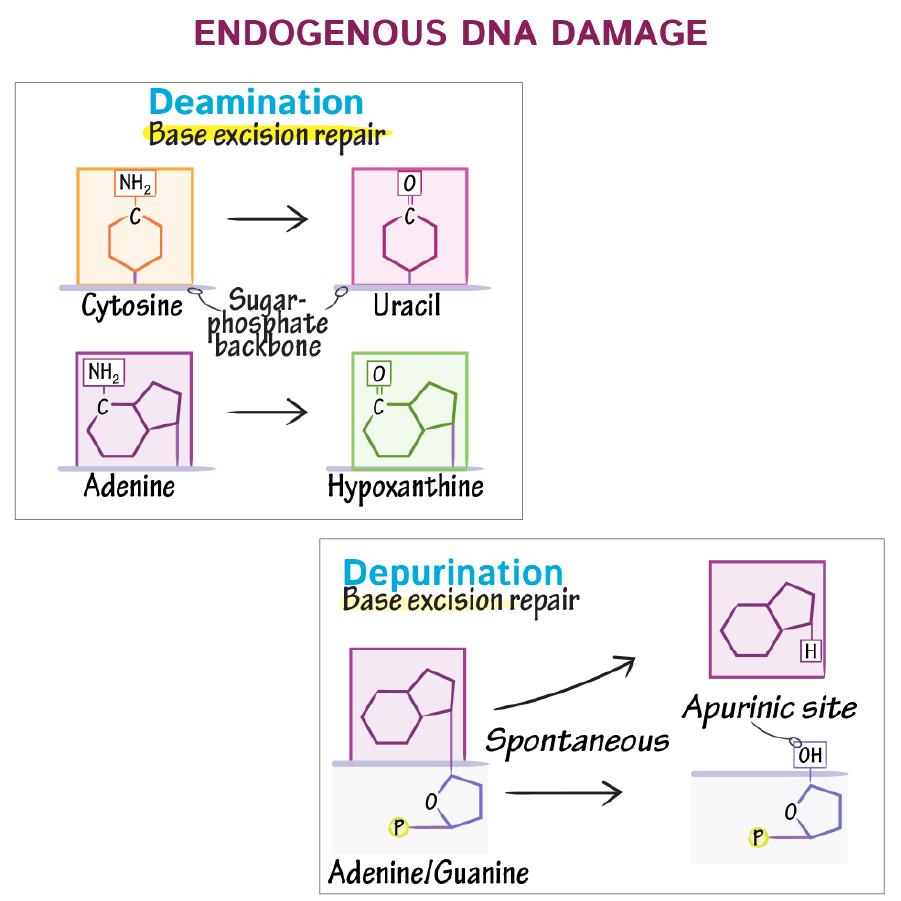
Spontaneous DNA damage caused by water are hydrolytic reactions. What is deamination and depurination?
Deamination: In pyrimidine bases (C, T) Loss of amino group in the presence of water that converts the base such as cytosine into uracil
Depurination: Loss of purine bases (A, G) due to cleavage of bond between base and deoxyribose sugar, leaving an apurinic site in DNA, leading to random mutations which can cause genome instability or cell death
Describe how the structure of DNA supports the accuracy of DNA repair mechanisms
Since DNA is double stranded and each strand is complementary to the other, the complementary strand can be used as a template to restore the correct nucleotide sequence
List and describe large mutations
Gene duplication - entire gene copied
Inversion - change orientation of gene (becomes “flipped”)
Genome duplication
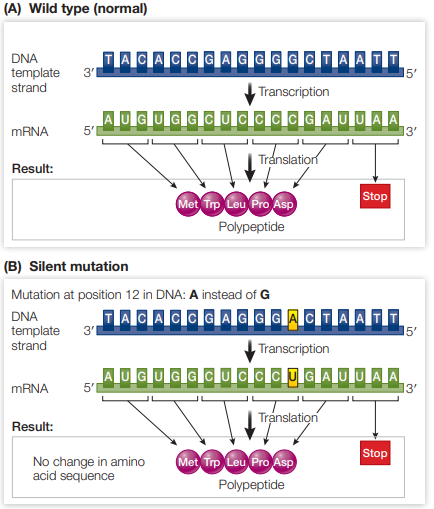
What is the outcome of a synonymous (or silent) mutation in a gene?
Change in nucleotide to another codon triplet does not affect the amino acid it codes for due to the degeneracy of the genetic code
Gene would encode for a functional protein (same as gene without synonymous mutation)
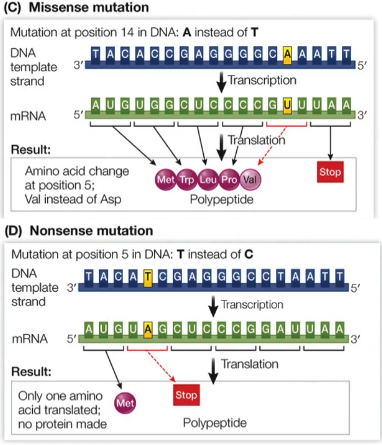
What is the difference between the non-synonymous mutations?
Non-synonymous mutations:
Missense mutations - substitution of nucleotide in DNA sequence results in a sense codon triplet encoding for a different amino acid, may result in gain of function or loss of function in the synthesised protein
Nonsense mutations - substitution of nucleotide in DNA sequence results in codon triplet encoding for a stop codon, results in a shortened protein
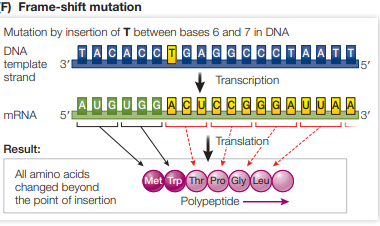
Explain how frameshift mutations affect a gene and mRNA that it encodes.
Frameshift mutations are when a base pair is inserted or deleted from a DNA sequence.
This alters the mRNA reading frame (the consecutive triplets), causing a different amino acid sequence
Therefore, the resultant protein after translation is often non-functional.
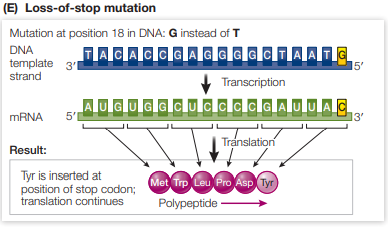
What is the effect of a loss of stop mutation?
The base pair substitution results in a stop codon being changed to a sense codon which encodes for an amino acid, resulting in more amino acids being added to the end of the chain
Effect is dependent on how many amino acids are added and how important that part of the polypeptide is to function
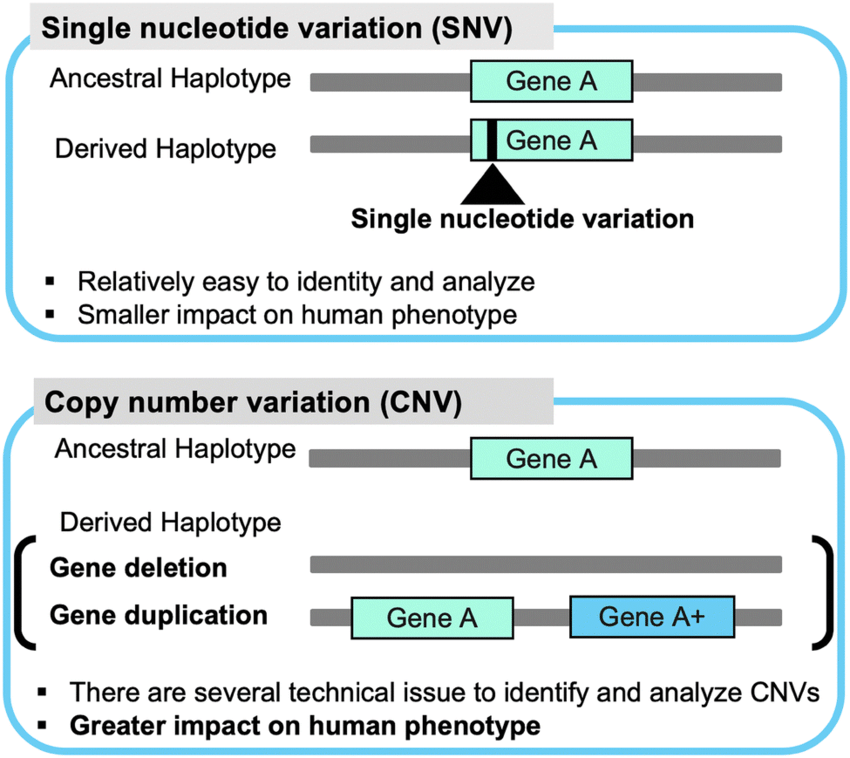
Explain the difference between copy number variation (CNV) mutations and single nucleotide polymorphism (SNP) mutations.
CNV mutations are large mutations that cover >1kbp, these can be duplicated through duplication and insertion
SNP mutations are single nucleotide substitutions that occur in >1% of the population
What are alleles?
Variations of the same gene
Diploid organisms inherit an allele of each gene from each parent
Explain the difference between hemizygous, homozygous and heterozygous genotypes
Hemizygous - gene has one allele (A)
Homozygous - gene has two of the same alleles (ex: A/A)
Heterozygous - gene has two different alleles (ex: A/a)
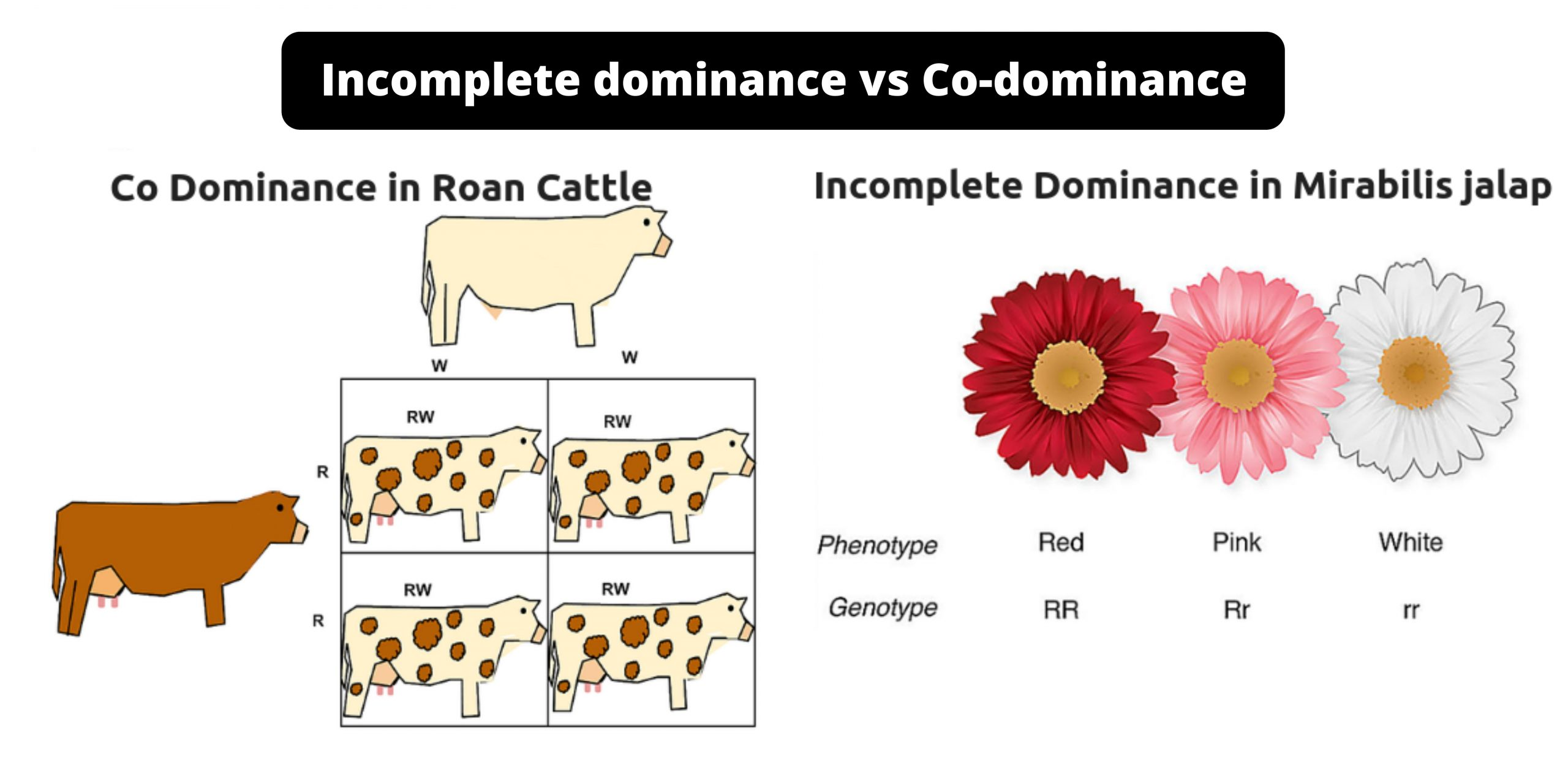
Define incomplete dominance and explain how it differs to co-dominant phenotypes.
Incomplete dominance is when a heterozygote shows a phenotype that is intermediate between those of the two homozygotes
ex: Rose colour gene, alleles Red (RR), White (rr), cross between the two homozygotes makes (Rr) which is pink due to incomplete dominance
Codominance is when the two alleles which produce different phenotypes are simultaneously expressed in the heterozygote
ex: codominant AB blood type makes both A and B antigens and does not make antibodies against either antigen