DNA Replication
1/110
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
111 Terms
What is the primary biological function of homologous recombination in bacteria?
To repair double-strand breaks (DSBs) using a homologous DNA template, preserving genomic integrity.
What are common sources of double-strand breaks in bacterial cells?
Ionizing radiation, replication fork collapse, and synthetic breaks induced by bacteriophages during infection.
What is the first protein complex involved in bacterial homologous recombination?
The RecBCD complex.
What are the helicase activities within the RecBCD complex and their directionality?
RecB is a 3’→5’ helicase and RecD is a 5’→3’ helicase.
What happens when RecBCD encounters a Chi site during resection?
It undergoes a conformational change, which increases degradation of the 5’ strand and promotes formation of a 3’ single-stranded overhang.
What is the role of the 3’ overhang generated by RecBCD?
It serves as a substrate for RecA loading and initiates strand invasion.
How does RecA bind to DNA, and what is its structure during this process?
RecA binds as a right-handed helical filament using ATP, with each monomer interacting with 3 nucleotides on the ssDNA.
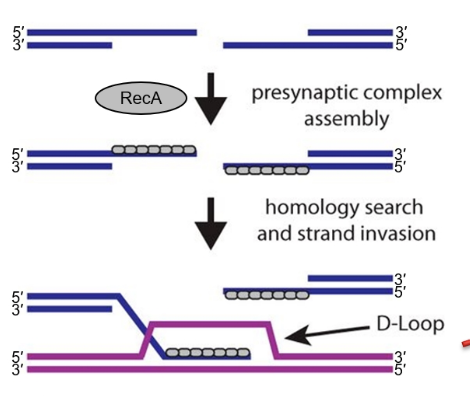
What triggers successful strand invasion by RecA?
Stable pairing of the invading strand with a homologous sequence of ~20 base pairs.
What is the name of the three-stranded structure formed during strand invasion?
D-loop (Displacement loop).
What protein binds to the Holliday junction during bacterial recombination?
RuvA tetramer binds to the junction.
What is the function of RuvB in recombination?
RuvB is an ATP-dependent hexamer that promotes branch migration of the Holliday junction.
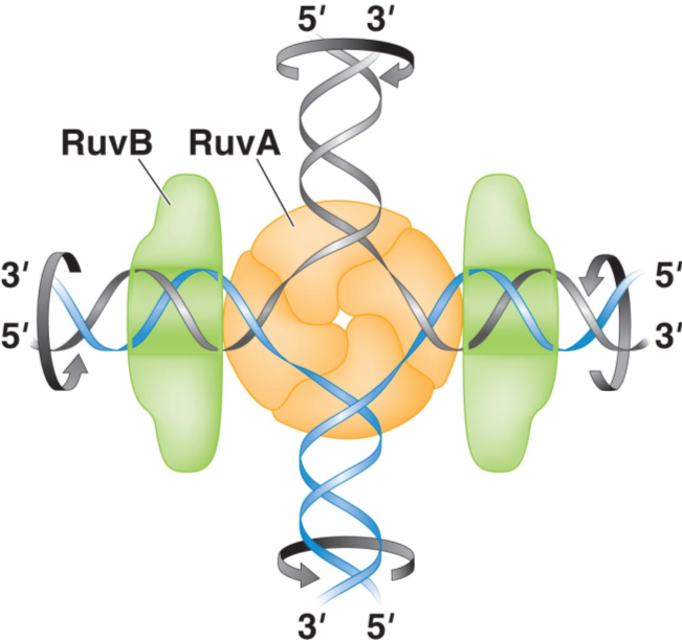
How is DNA synthesis coupled to strand invasion and branch migration?
DNA polymerase extends the 3’ end of the invading strand, copying DNA from the homologous template.
What structure forms after the extended strand reanneals to the original chromosome?
A double Holliday junction.
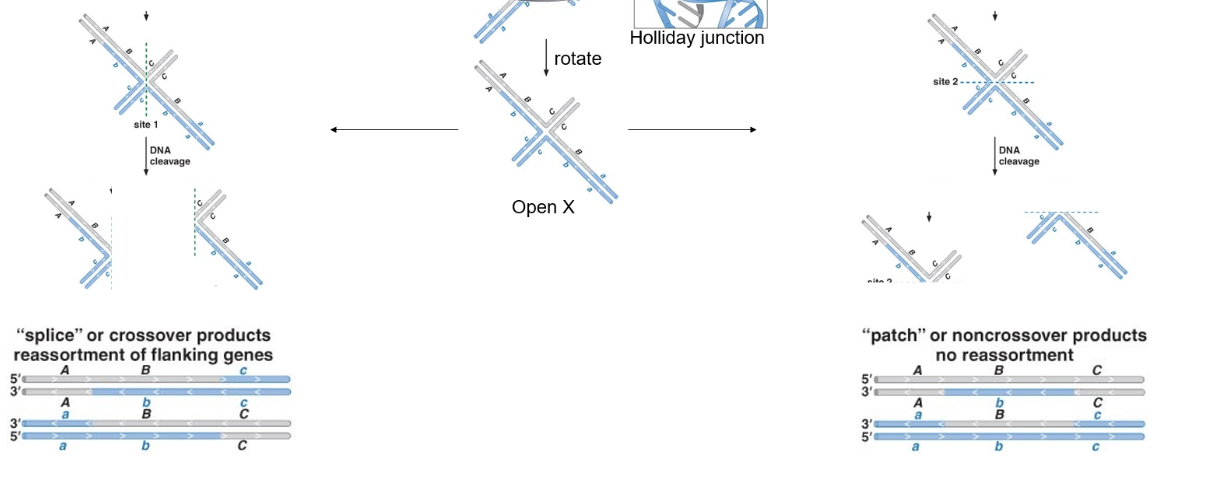
What is the role of RuvC in recombination?
RuvC is a resolvase that cleaves Holliday junctions to resolve recombinant products into either patches or splices.
What determines whether the recombination products are “patch” or “splice”?
The orientation and position of RuvC cleavage across the junction.
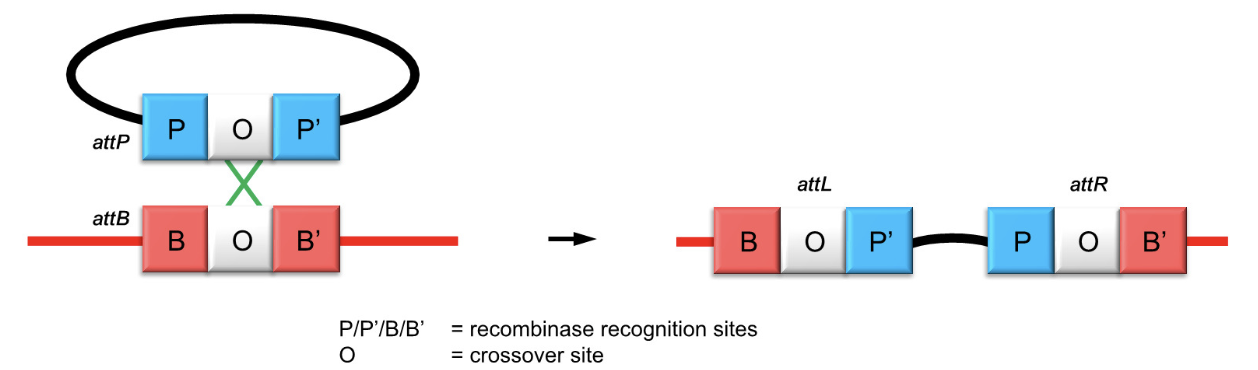
How is homologous recombination used by lysogenic bacteriophages?
Phages use site-specific recombination between phage attachment site (attP) and bacterial site (attB) using a tyrosine recombinase to integrate their genome.
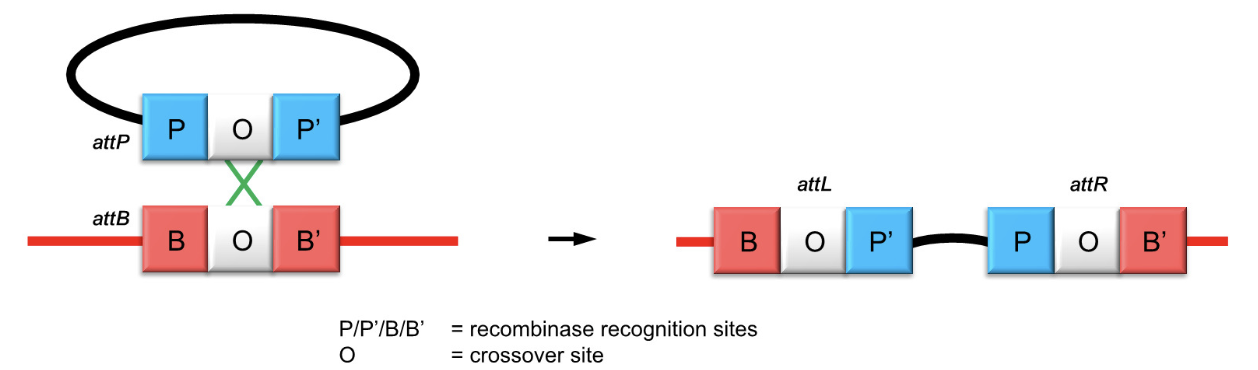
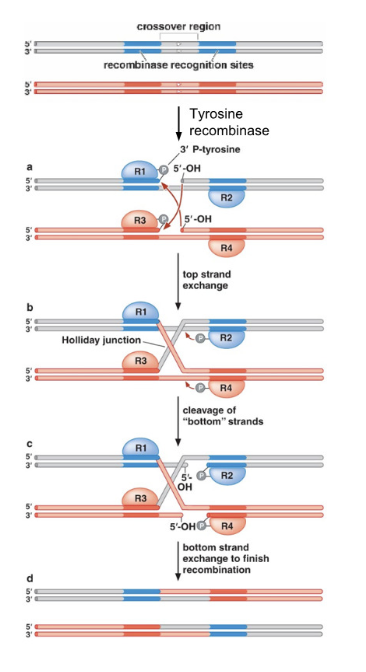
What enzymatic mechanism does the phage tyrosine recombinase use?
It cleaves DNA using a tyrosine residue to form covalent protein-DNA intermediates and facilitates strand exchange.
What role does recombination play in meiosis?
It facilitates crossing over between homologous chromosomes to increase genetic diversity.
Which enzyme initiates recombination in meiosis?
Spo11 introduces programmed DSBs at specific hotspots.
How is Spo11 removed, and what protein complex initiates resection in meiosis?
The MRX complex (Mre11-Rad50-Xrs2) and Sae2 remove Spo11 and initiate resection.
Which protein promotes strand invasion during meiotic recombination?
Dmc1, a meiosis-specific homolog of RecA.
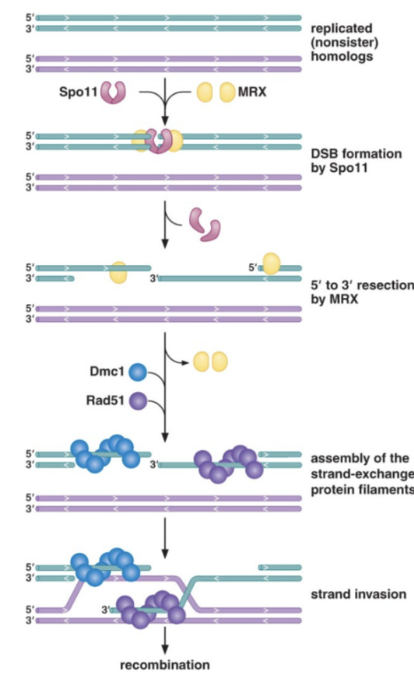
What is the role of RPA in eukaryotic recombination?
RPA binds and stabilizes ssDNA after resection.
How is Rad51 filament formed on ssDNA during eukaryotic recombination?
Rad52 displaces RPA and recruits Rad51 to form a nucleoprotein filament, stabilized by Rad55 and Rad57.
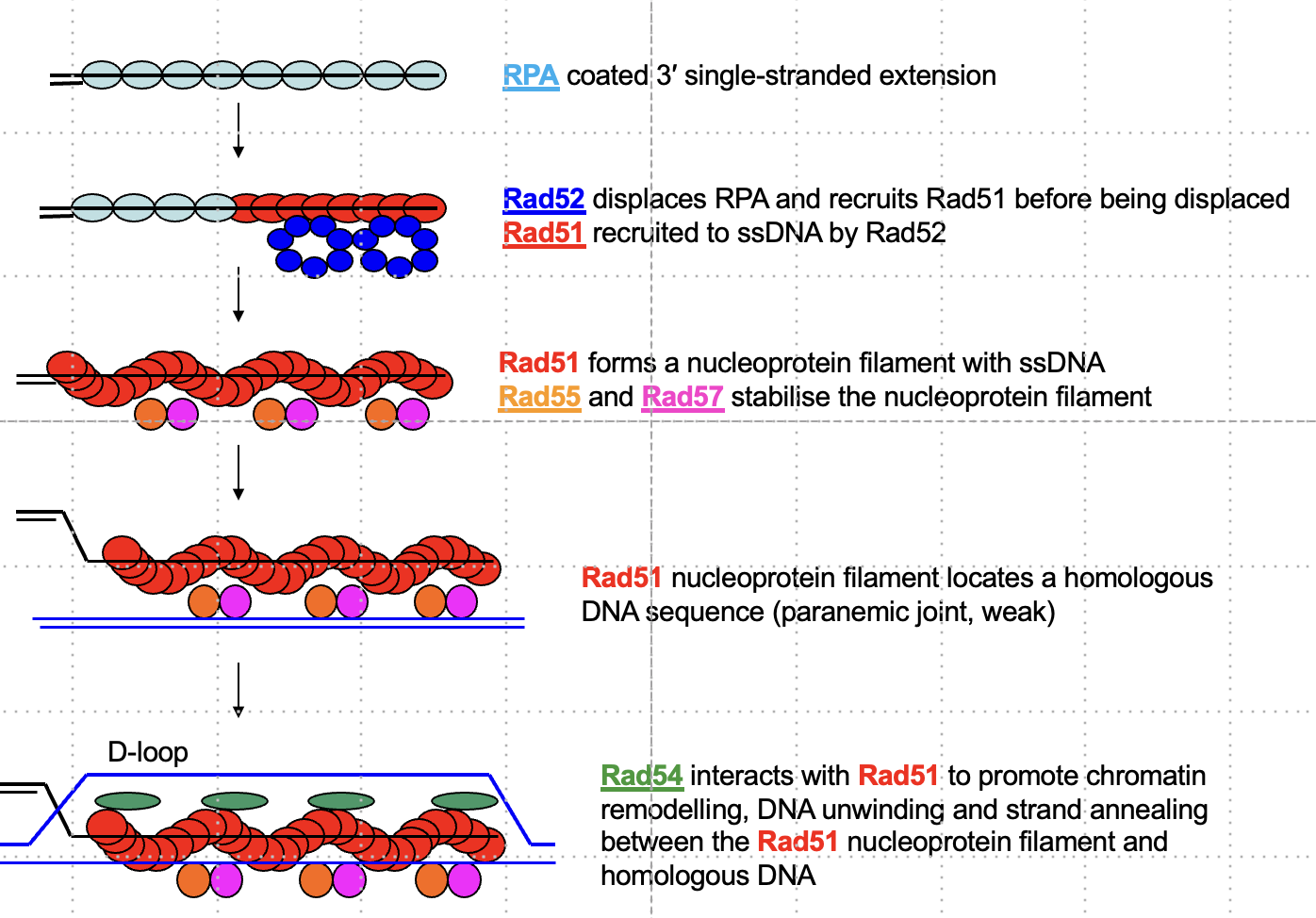
Why is Rad54 necessary for strand invasion in eukaryotes but not bacteria?
Rad54 remodels chromatin to expose homologous regions and aids in DNA unwinding, which is not needed in naked bacterial DNA.
How is the resolution of recombination intermediates similar in prokaryotes and eukaryotes?
Both resolve double Holliday junctions through cleavage or dissolution pathways to produce crossover or non-crossover products.
How does the protein toolkit differ between bacterial and eukaryotic homologous recombination?
Eukaryotes use a more complex set of proteins including MRX, Sae2, RPA, Rad52, Rad51, Rad55/57, and Rad54 due to chromatin structure and regulatory needs.
What signals RecBCD to change its activity during homologous recombination?
Encountering a Chi site.
What does the Chi site do to RecBCD function?
It causes a conformational change in RecBCD that enhances degradation of the 5’ strand, promoting 3’ overhang formation.
What are the specific helicase directions of RecB and RecD?
RecB is a 3’ to 5’ helicase; RecD is a 5’ to 3’ helicase.
What is the structure and binding mode of RecA on single-stranded DNA?
RecA forms a right-handed helical filament; each monomer binds to 3 nucleotides in an ATP-dependent manner.
What DNA structure forms during strand invasion?
A displacement loop (D-loop), where the invading strand pairs with a homologous sequence.
What is the function of RuvA in homologous recombination?
RuvA binds to Holliday junctions and stabilizes the structure by targeting all four strands.
How does RuvB function in homologous recombination?
RuvB is an ATP-dependent hexametric motor that drives branch migration of the Holliday junction.
What role does DNA polymerase play after strand invasion?
It extends the 3’ end of the invading strand before rejoining to the 5’ end.
What is the function of RuvC?
RuvC is a nuclease that resolves Holliday junctions by cutting across the junction.
What are the two possible outcomes of Holliday junction resolution?
"Patch" products (non-crossover) or "splice" products (crossover with flanking exchange).
How do bacteriophages use homologous recombination during lysogeny?
They use site-specific recombination between attP (phage) and attB (bacterial) sites via tyrosine recombinase to integrate into the host genome.
What enzyme initiates double-strand breaks during meiotic recombination?
Spo11.
What complex removes Spo11 and carries out resection in meiosis?
The MRX complex.
What is the eukaryotic homolog of RecA?
Rad51.
What protein promotes Rad51 filament formation on single-stranded DNA?
Rad52.
What proteins stabilize the Rad51 filament?
Rad55 and Rad57.
What is the function of Rad54 in eukaryotic homologous recombination?
It remodels chromatin and unwinds DNA to promote homology search and strand invasion.
How is the process of homologous recombination in eukaryotes more complex?
It involves additional proteins for resection, filament stabilization, and chromatin remodeling compared to bacteria.
e does bacterial chromosome replication initiate?
At the OriC, the origin of bidirectional replication.
What are DnaA boxes and trios?
Sequences at OriC where DnaA binds; boxes bind DnaA oligomers, and trios are triplet motifs (GAT) targeted for DNA unwinding.
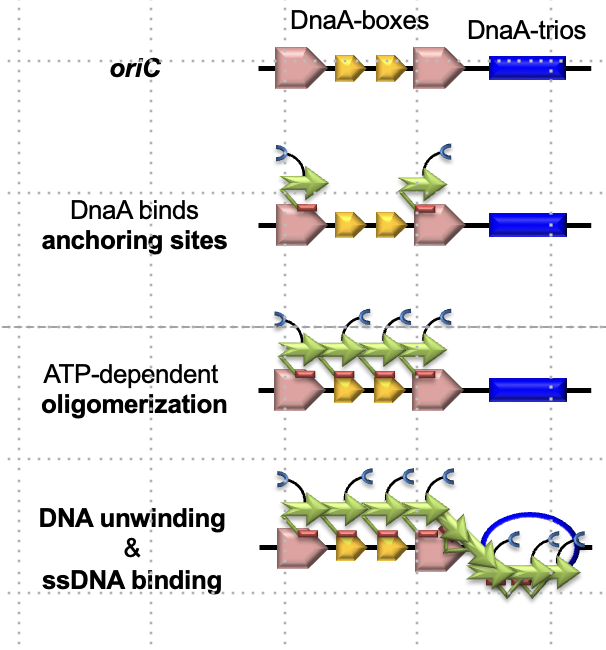
What is the role of DnaA in replication initiation?
It binds to DnaA boxes, oligomerizes using ATP, and unwinds DNA at trios to initiate strand separation.
How does DnaA specifically unwind DNA at the trios?
By capturing the G and A bases in the GTA sequence to separate the strands.
What protein loads DnaB helicase onto DNA?
DnaC, which uses ATP to load DnaB and then dissociates.
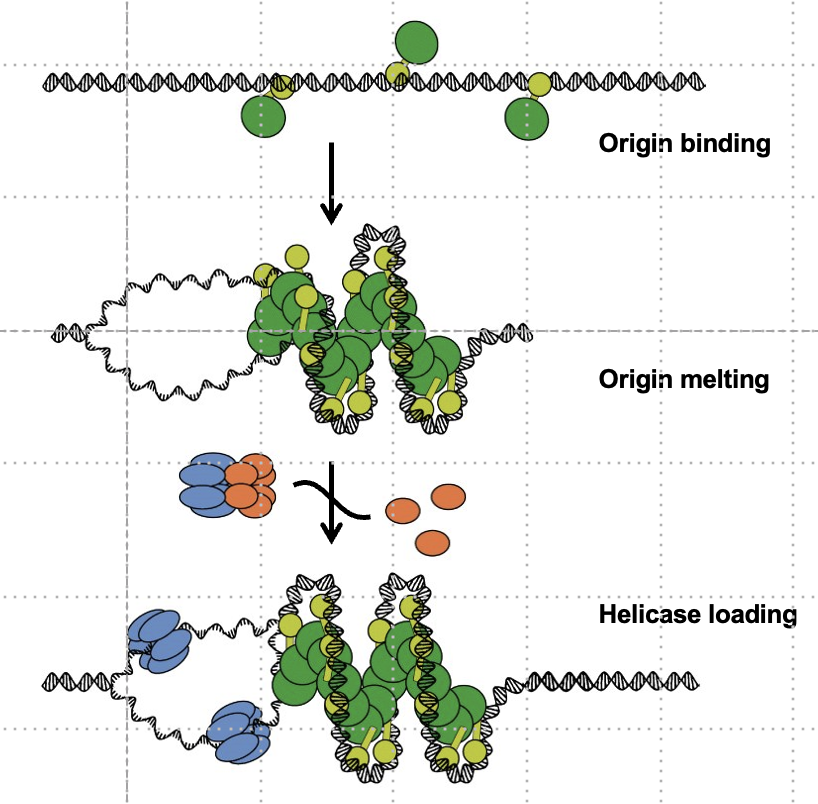
In which direction does DnaB helicase move, and what is its role?
5’ to 3’ on the lagging strand; it unwinds DNA in an ATP-dependent manner.
What is the function of single-strand binding protein (SSB)?
It stabilizes single-stranded DNA, prevents secondary structures, and protects from degradation.
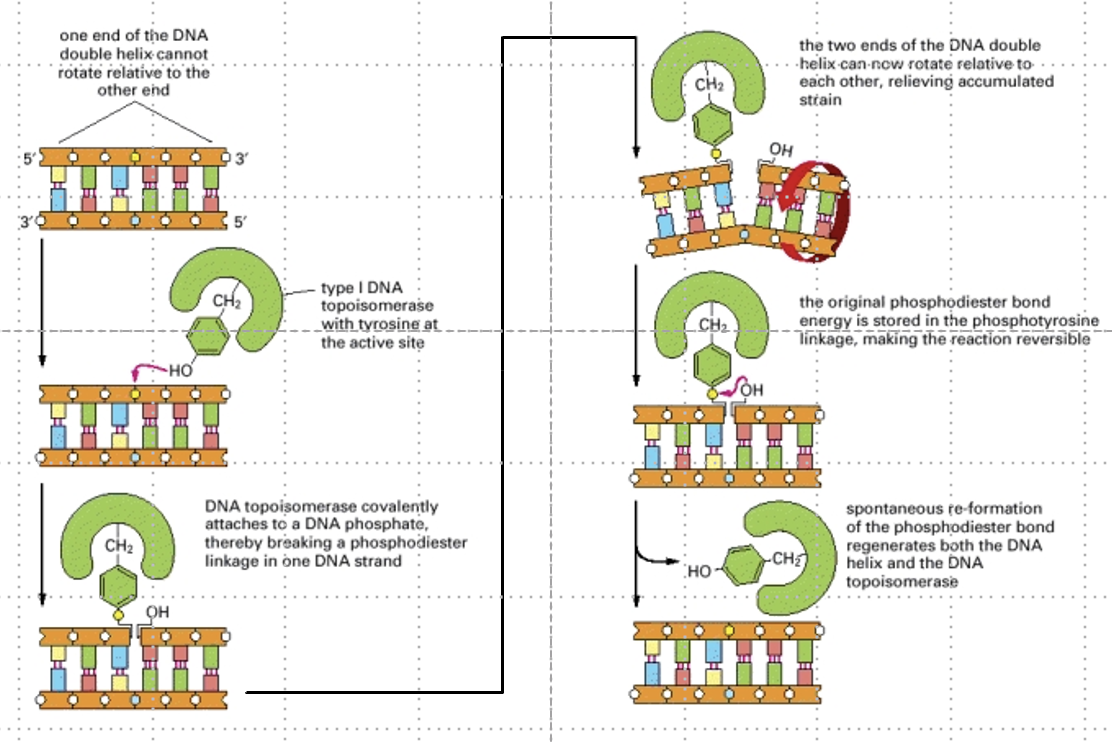
Why is topoisomerase required during replication?
To relieve supercoiling ahead of the replication fork by cleaving and rotating one DNA strand.
How does topoisomerase relieve supercoiling?
It uses a tyrosine residue to nick one strand and allow it to rotate, releasing tension.
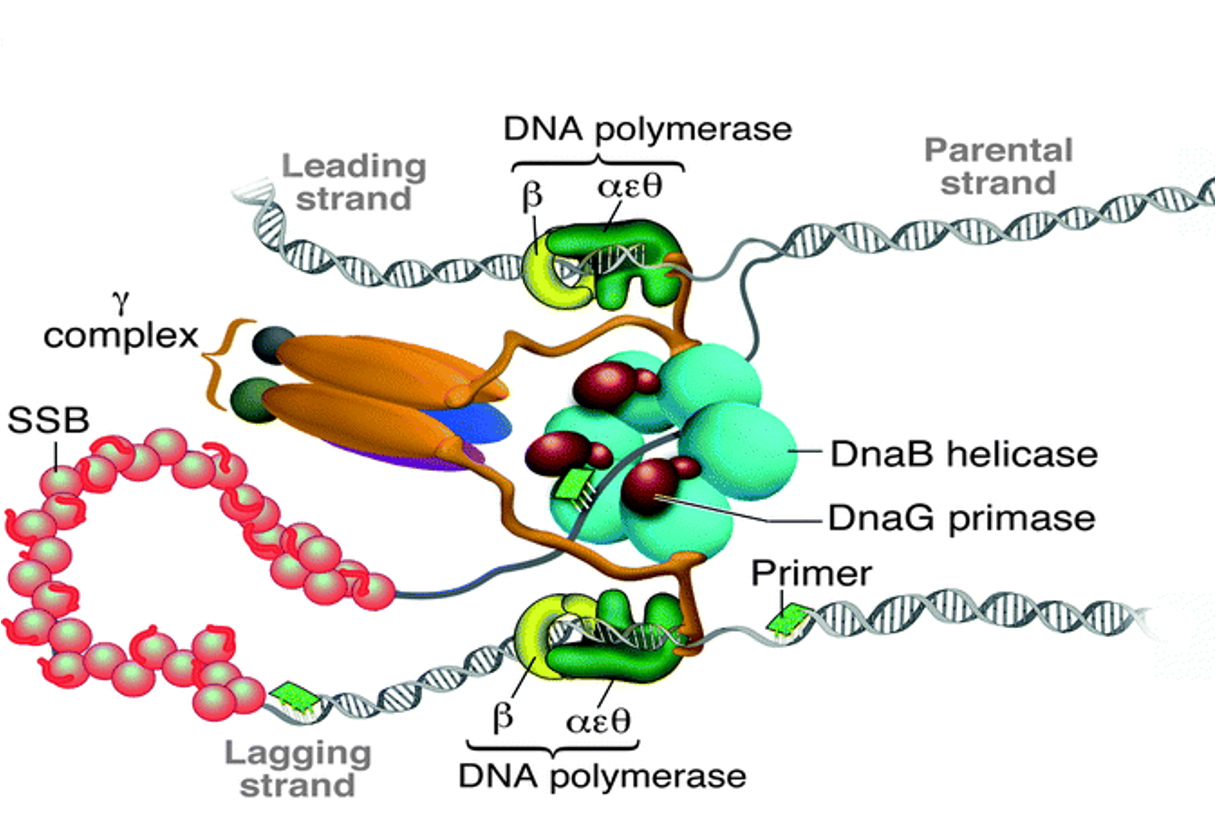
What is the function of DnaG?
DnaG is primase that synthesizes a short RNA primer (~5 nucleotides) for DNA polymerase III to begin synthesis.
What protein recruits the clamp loader and DNA Pol III?
DnaB helicase.
What is the role of the clamp loader?
It uses ATP to load the β clamp, which tethers DNA polymerase III for processivity.
What direction does DNA polymerase III synthesize in?
5’ to 3’.
How does DNA polymerase III interact with DNA during synthesis?
It grips DNA like a fist, with fingers binding dNTPs and the palm as the catalytic site; the thumb helps with DNA exit.
What role do magnesium ions play in the active site of DNA Pol III?
They stabilize pyrophosphate and deprotonate the 3’ OH for nucleophilic attack during phosphodiester bond formation.
How is the lagging strand synthesized despite its opposite directionality?
Via discontinuous replication using Okazaki fragments, with repeated priming by DnaG and synthesis by Pol III.
What is the role of DNA polymerase I on the lagging strand?
It replaces RNA primers with DNA and has 5’ to 3’ exonuclease activity.
How is the nick between Okazaki fragments sealed?
DNA ligase seals the nick using NAD+ or ATP to donate AMP and facilitate phosphodiester bond formation.
What is the trombone model of replication?
It describes how the lagging strand loops to allow both strands to synthesize in the same direction, coordinated by the β clamp.
What signals termination of replication in bacteria?
Ter sites that bind Tus protein, forming the Tus-ter complex.
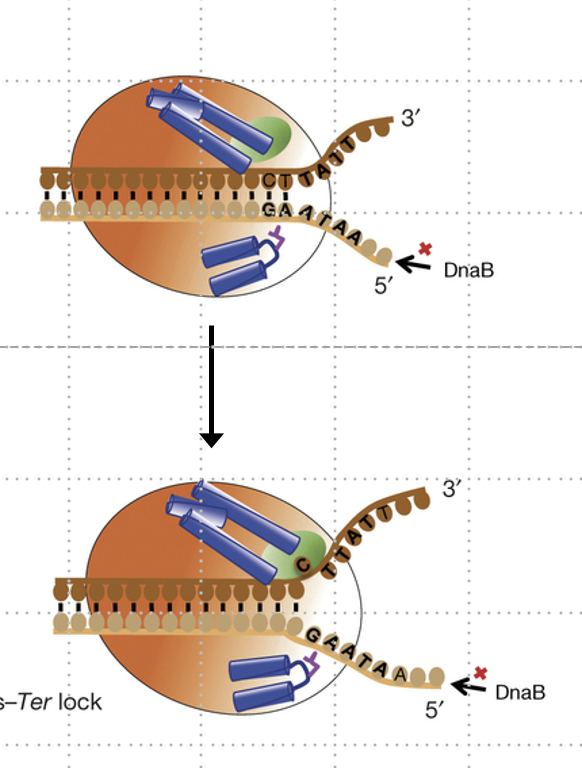
How does the Tus-ter complex regulate termination directionally?
It allows the fork to pass in one direction but arrests it in the other by unwinding a GC base pair into Tus’s binding pocket.
What is the structure of the DnaA oligomer, and how is it stabilized?
A right-handed helical filament formed in an ATP-dependent manner; each monomer binds a DnaA box.
Why is the bacterial chromosome’s circular structure significant in replication?
It necessitates bidirectional replication and specific termination sites to avoid over-replication and head-on replisome collisions.
hat type of enzyme is DnaA, and what two functions does it have?
It is a multidomain protein with helicase activity and AAA+ ATPase function.
How does the beta clamp affect DNA polymerase III activity?
It increases processivity by tethering the polymerase to DNA.
What experimental method led to the discovery of Okazaki fragments?
A pulse-chase labeling experiment conducted by the Okazakis in 1968.
What is required for DNA ligase to function in sealing nicks?
Either NAD⁺ or ATP as an AMP donor.
Why must primase (DnaG) be recruited repeatedly on the lagging strand?
Because DNA synthesis is discontinuous, requiring multiple RNA primers.
How does the DNA polymerase III "fist" structure relate to its function?
Fingers bind dNTPs, the palm catalyzes bond formation, and the thumb holds the duplex for stability.
What structural difference in chromosomes necessitates multiple origins of replication in eukaryotes?
Eukaryotes have longer, linear chromosomes, requiring multiple bidirectional origins.
What sequences mark replication origins in yeast, and what is their key characteristic?
Autonomously replicating sequences (ARS), which are rich in AT nucleotides.
What is the eukaryotic functional homolog of bacterial DnaA?
The Origin Recognition Complex (ORC).
What helicase complex is used in eukaryotes, and how is it loaded?
MCM2-7 helicase, loaded by Cdc6 and Cdt1 as a double hexamer around dsDNA.
What activates the MCM helicase into the CMG complex?
DDK phosphorylates MCM to allow Cdc45 binding; CDKs then phosphorylate Sld2/3 to recruit GINS.
What does the CMG complex consist of?
Cdc45, MCM2-7, and GINS.
What is the role of MCM10 in helicase activation?
MCM10 activates MCM2-7 helicase for 5’ to 3’ unwinding on the leading strand.
How does the MCM helicase manipulate DNA during activation?
It induces a left-handed kink in DNA, opens the MCM2-5 gate, and captures one strand via MCM10.
How does MCM helicase accommodate bidirectional replication?
Once activated, the helicase units can pass each other, enabling replication forks to proceed in opposite directions.
Which bacterial replication components have conserved analogs in eukaryotes?
Primase, sliding clamp (PCNA), and DNA polymerases.
What replication-specific challenge is posed by eukaryotic chromatin?
The presence of histones and the need to faithfully re-deposit them with correct epigenetic marks.
How are histones transferred during replication in eukaryotes?
MCM2 binds and displaces histones from the parental strand; they are redistributed via Dpb3-4 (to leading) or directly to lagging strand.
What ensures chromatin reassembly on strands that did not inherit parental histones?
Chromatin Assembly Factor 1 (CAF1) deposits newly synthesized histones.
What prevents eukaryotic origins from firing more than once per cell cycle?
Licensing occurs only during G1 via ORC, Cdc6, and Cdt1; once S phase begins, CDKs inactivate these factors to prevent re-licensing.
What are early- and late-firing origins, and how are they regulated?
Not all replication origins fire simultaneously; timing is regulated by chromatin structure, local histone modifications, and availability of initiation factors.
Which eukaryotic DNA polymerases are responsible for lagging and leading strand synthesis?
Pol ε synthesizes the leading strand; Pol δ synthesizes the lagging strand; Pol α/primase initiates both strands.
How is the RNA primer removed and replaced during eukaryotic Okazaki fragment maturation?
Pol δ displaces the RNA primer as a flap, which is cleaved by FEN1, and DNA ligase I seals the resulting nick.
What is a replication factory?
A replication factory is a nuclear subdomain where multiple replication forks are anchored and coordinated during S phase.
How do CDKs regulate DNA replication during the cell cycle?
CDKs activate origin firing by phosphorylating initiation factors (e.g., Sld2/Sld3) during S phase and inhibit new licensing until the next G1.
Why do eukaryotes require telomerase for replication?
Linear chromosomes leave a 3' overhang after lagging strand synthesis, which telomerase extends using an RNA template to avoid progressive shortening.
Which complex is responsible for loading MCM helicase onto DNA?
The pre-replicative complex (pre-RC) formed in G1 includes ORC, Cdc6, and Cdt1, which load MCM2-7 as a double hexamer.
What activates the MCM helicase in eukaryotes?
DDK phosphorylates MCM to allow Cdc45 binding, and CDKs phosphorylate Sld2/Sld3 to recruit GINS, forming the CMG complex.
What protein helps recycle histones during eukaryotic replication?
MCM2 binds parental histones and deposits them on the lagging strand or via Dpb3/4 on the leading strand; CAF1 deposits new histones.
What is the bacterial replisome?
A multi-protein complex responsible for coordinating DNA unwinding and synthesis during replication.
What is the function of DnaB in the bacterial replisome?
DnaB is a 5' to 3' helicase that unwinds DNA ahead of the replication fork on the lagging strand template, using ATP.
How does DnaB recruit primase and what does primase do?
DnaB recruits primase (DnaG) to synthesize a short RNA primer (~5 nucleotides) providing a 3'-OH for DNA Pol III.
How does DNA Pol III synthesize DNA?
It uses an open-palm grip to position the template and incoming dNTPs, with two Mg²⁺ ions catalyzing the phosphodiester bond.