microbial ecology - HG2
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
16 Terms
Microenvironments
large changes in chemistry over very small distances
nutrient access limited by diffusion (like oxygen for ex)
natural vs lab growth rates
microbes have intermittent exposure to nutrients —> feat or famine lifestyle
e.coli divide in lab way faster than in the human gut naturally
this is bc resources and conditions, non-uniform distribution, microbes have to grow in mixed communities
lowk obvi!
Enrichment Culture
culture-DEPENDENT
whos out there and what r they doing
isolate individual microbes/communities w particular metabolic abilities
Steps:
inoculum from habitat w unknown # of organisms
enrichment conditions promote growth of specific microbes
positive result = organism w selected property was present (DK HOW MUCH tho)
negative = inconclusive (cant prove it DOESN’T exist)
Workflow:
medium w select nutrients —> sample w hella organisms —> organism of interest multiplies —> BACK dilute to first tube till enough to plate —> pure culture of organism of interest
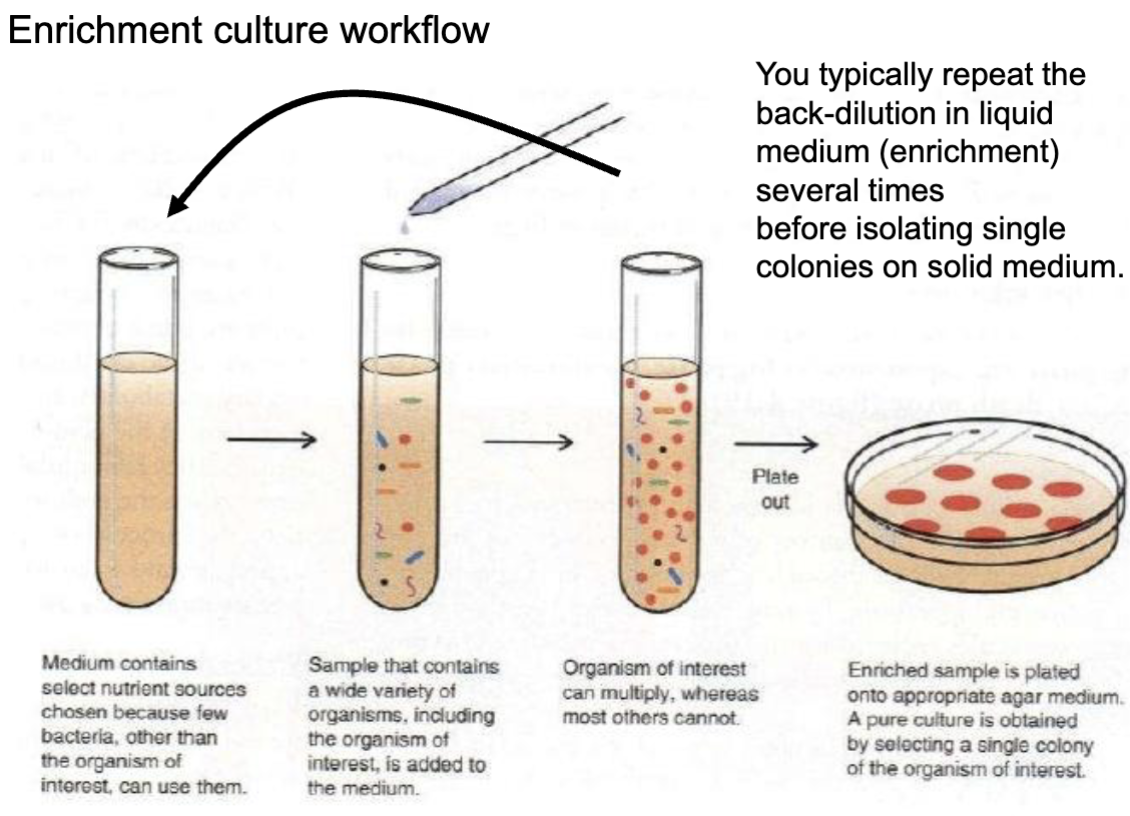
Enrichment Bias
liquid enrichment cultures —> rapidly growing microbes that perform the selected metabolism dominate
even if they arent the most abundant microbes in the OG environment that can do that type of metabolism
fast > amount
Lab environments let the slow ones grow faster
we end up indirectly selecting for the fastest growing bacterium in the sample that can do that metabolism.
Avoiding enrichment bias
Dilute initial inocolum and grow multiple independent enrichment cultures —> rare but fast “weed species” don’t take over every culture
Cell sorting —> separate individual cells of inoculum into wells of 96 well plates —> microbes in inoculum not in competition with eachother for nutrients
can grow at a range of time to find the fast vs slow one
Enrichment for bacteria that degrade plastic waste
PET = polyethylene terephthalate = pollutant in landfills/ocean
Bacteria can use PET as a carbon source, they make PETase and MHETase that can break it down
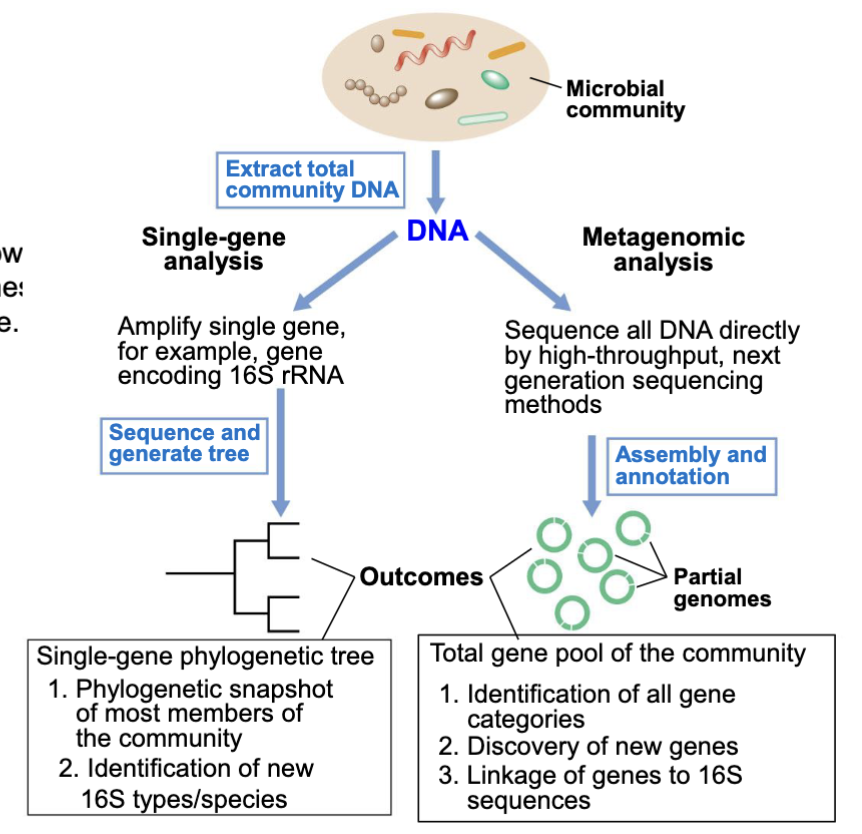
Single Gene Analysis
(culture independent)
looks at diversity of microbes in a sample (who’s out there)
isolate genomic DNA from environment —>
amplify selected genes from DNA sample (16s rRNA) —>
obtain sequences —>
BLAST sequence to ID known organisms w 16S sequence matches —>
create phylogenetic tree of all sequences present including ones that don’t correspond to existing species
most abundant is usually ones that haven’t been cultured in the lab → unknown
Perturbed microbial community changing over time
deepwater horizon spill in gulf of mexico —> they did single gene analysis of bacterial 16s rRNA to characterize changes in the community after
organismal diversity decreased, got dominated by 2 types that consume oil as a carbon source
community became diverse again after oil was gone
Metagenomics vs Single gene analysis
metagenomics (culture independent):
not only ID nif genes, but also know what 16s sequence is on the same DNA fragment
can answer question “who in this habitat can fix N2”
Single gene is.. well.. for a single gene!
Metatranscriptomics
gives a better idea of what microbes were actually doing when sample was collected
total DNA and RNA collected
RNA rev trans —> DNA —> sequenced —> genes transcribed in environment revealed —> metagenome made —> transcripts associated with contiguous 16S rRNA
lets us say than an organism w/particular 16S was or wasn’t transcribing particular genes at the time of sample collection
Q: what were they doing!
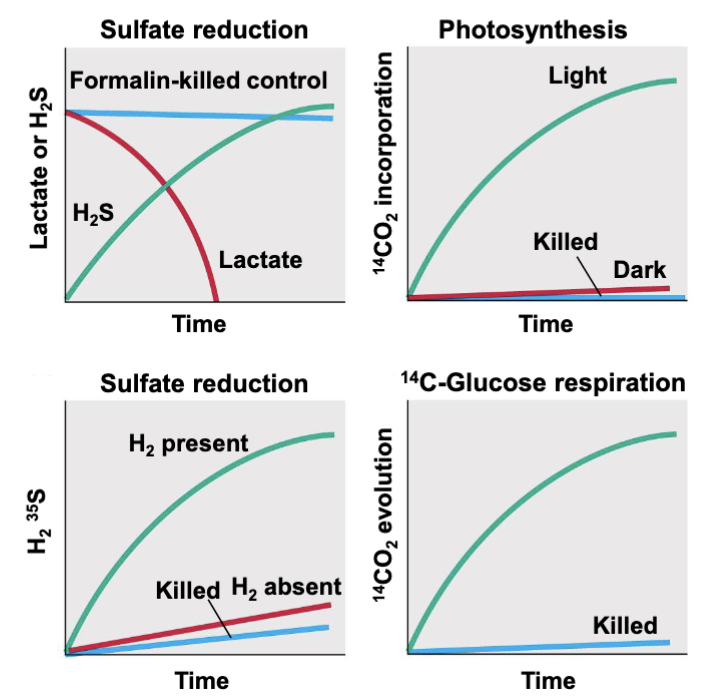
Chemical assays
reveals processes happening in an environmental sample
need a killed control to account for abiotic processes that could prouce the measured compounds
assays give rates of reactions occurring in samples from specific environments
DONT tell u WHAT organism is present or WHO IS DOING IT
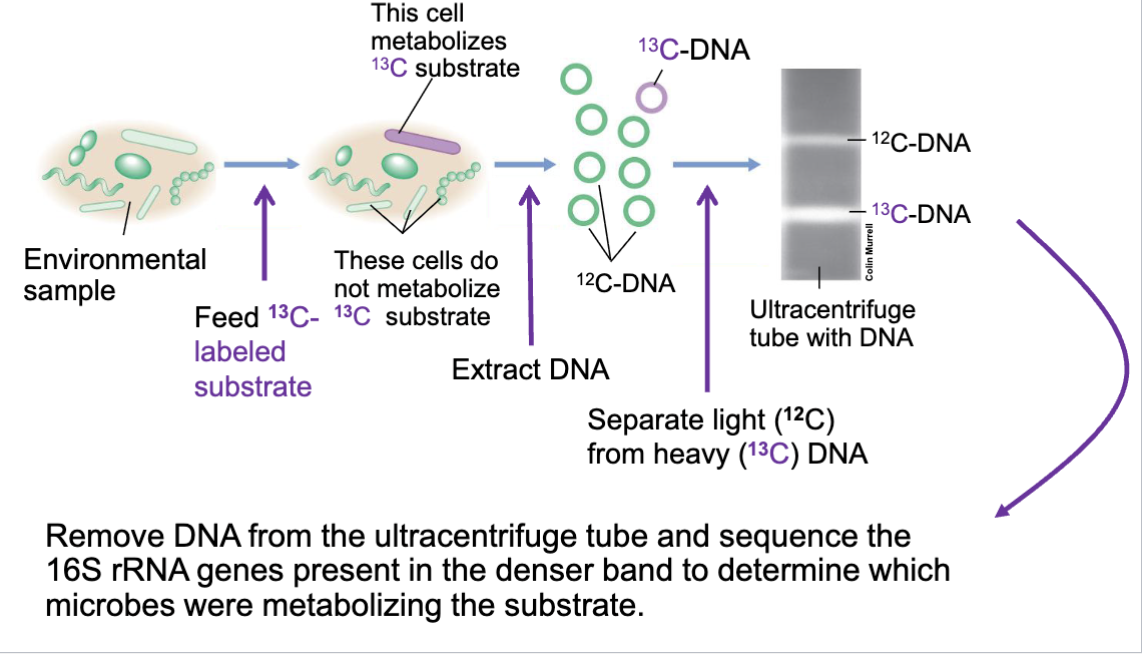
Stable Isotope Probing
assosciates reactions w organisms b4 performing them
measures use of substrates containing C or N + version of substrate with stable isotope needed (C 13 or N 15)
who is eating what.
KEY Qs
What are the methods and limitations of techniques
to assess which microbes are present in various habitats?
What can a metagenome (potentially) tell you that single-
gene community analysis can’t?
What are the methods and limitations of techniques to
determine what reactions are occurring in habitats?
What methods can reveal (or at least suggest) which
organism in a habitat is performing a particular reaction?
What is enrichment bias, and what methods are used
to avoid it?
any surface exposed to nutrients will be colonized
surfaces = nutrient, protection, means of staying in a favorable habitat vs getting washed away
abiotic surfaces: glass/rocks
organic surfaces: particles of decaying plants
other organisms as surfaces: plant roots, insects, each other
microcolonies = clusters of few cells that develop from a single attached cell
surface colonization can be sparse, detectable via microscope or hella dense
biofilms on surfaces can become microbial mats (cm thick) w diff types of microbes living in diff layers (tops of stromatolites)
Biofilms
predominant mode of bacterial growth in natural environments
group of bacteria enclosed in adhesive/self produced matrix
made up of exopolysaccharides (EPS), proteins, nucleic acids
adheres to abiotic or living things or free floats (flocs) in water
bacteria living in biofilms are in physiologically different states than if they were free
they can have diff growth rates, transcriptional profiles
they can hav enhanced tolerance toward antibiotics and enanced interactions w other microbes
environmental biofilms are usually multispecies communities, studies have been done on single tho.
Biofilms and human health / industry
biofilms grow on catheters, IV lines, and artificial joints in patients, fouling in fuel storage tanks, in water pipes: corrosion and water borne disease spread
biofilms help corrode submerged objects like boats/piers
dental plaque is also a biofilm w 100s of bacteria and archaea
organisms in biofilm can produce acid as a metabolic byproduct —> ex: tooth decay