11.3 - The activity of negative regulatory transcription factors - repressors (copy)
1/23
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
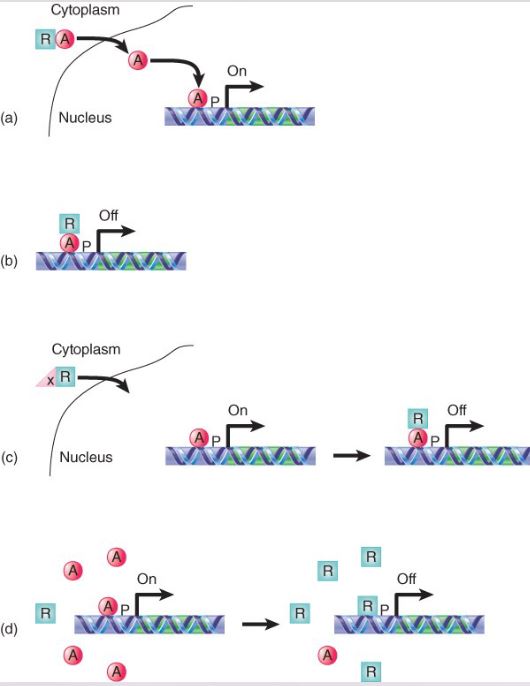
list the potential methods on how Repressors may control transcription by
a) sequestering an activator in the cytoplasm (keep activator from entering the nucleus)
b) by binding an activator and masking its activation domain
(c) by being held in the cytoplasm until it is needed
(d) by competing with an activator for a binding site
Mechanism of Action of Activators and Repressors (generally)
Some components of the transcriptional apparatus work by changing chromatin structure
Repression is achieved by affecting chromatin structure or by binding to and masking activators
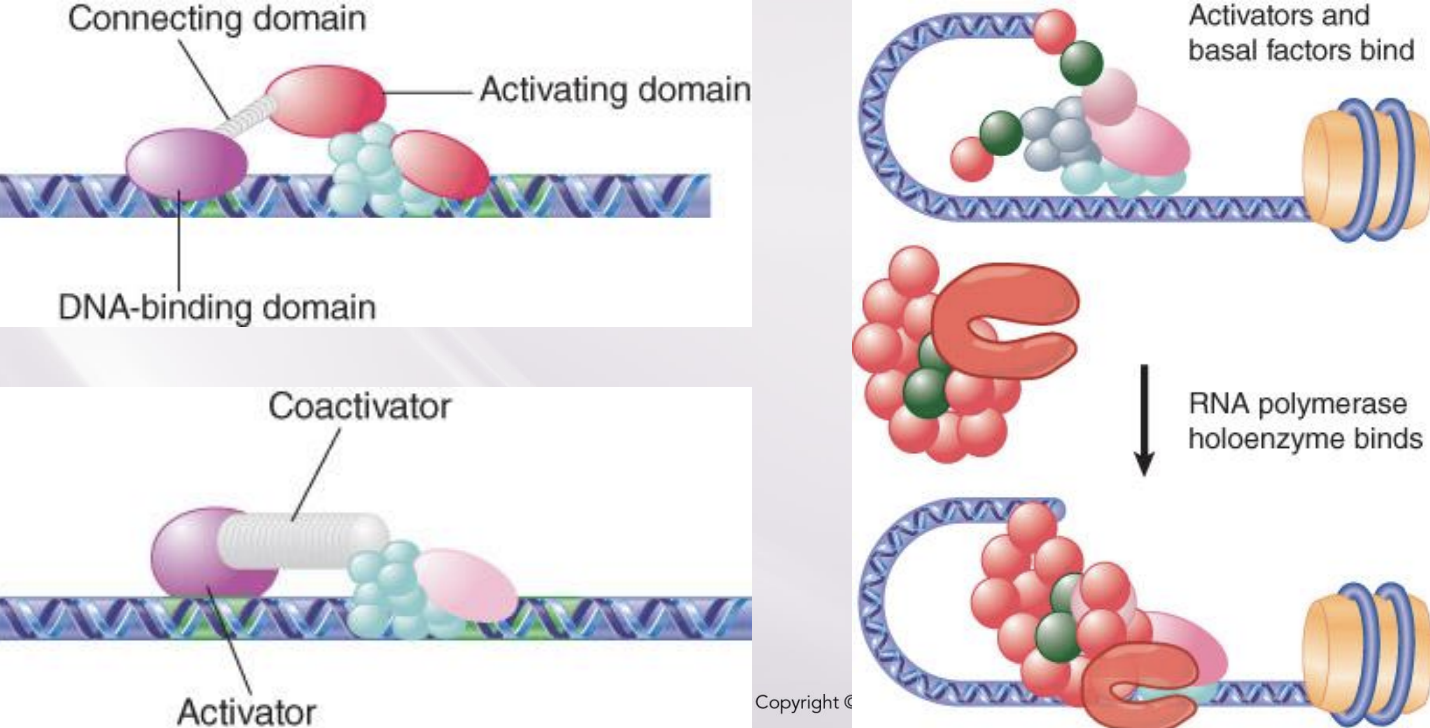
Activators and repressors often have DNA-binding and activating functions in independent domains of the protein.
Mediator complexes
associate with RNA polymerase and replace activators/co-activators and basal factors
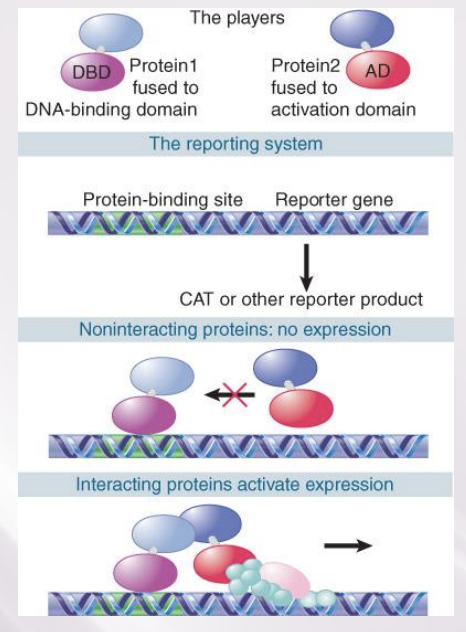
The Two-Hybrid Assay
purpose
how does it work?
Detects Protein–Protein Interactions
The two-hybrid assay works by requiring an interaction between two proteins, where one has a DNA- binding domain and the other has a transcription- activation domain
can’t find promoter by itself
interaction of 2 proteins brings activation domain to promoter
how do activators Interact with the Basal Apparatus
The principle that governs the function of all activators is that a DNA-binding domain determines specificity for the target promoter or enhancer.
The DNA-binding domain is responsible for localizing a transcription-activating domain in the proximity of the basal apparatus (RNA pol. and TFs)
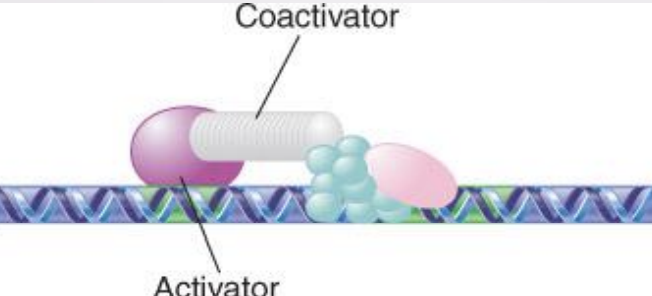
coactivator
An activator that does not have an activating domain may work by binding a coactivator that has an activating domain
Several factors in the basal apparatus are targets with which activators or coactivators interact
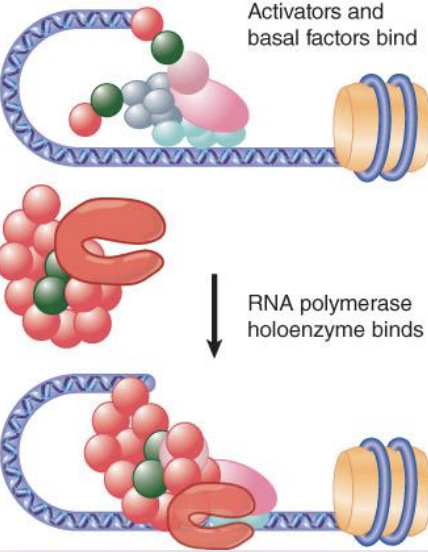
Mediator complexes
RNA polymerase may be associated with various alternative sets of transcription factors in the form of a holoenzyme complex.
Mediator complexes associate with RNA polymerase and replace activators/co-activators and basal factors
how are activators classified?
how do activators differ?
give an example of one type
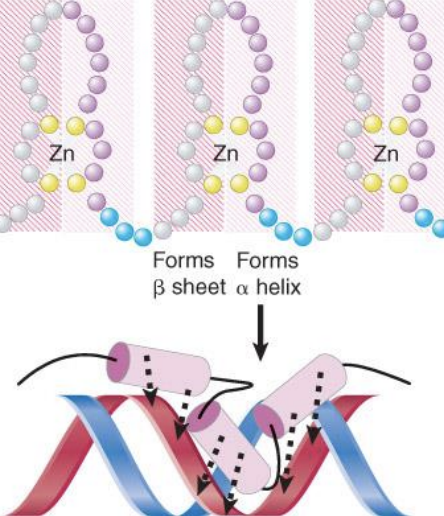
Activators are classified according to the type of DNA binding domain.
Members of the same group have sequence variations of a specific motif that confer specificity for individual DNA target sites
zinc finger-domain - DNA-binding motif that typifies a class of transcription factor
steroid receptor
transcription factors that are activated by binding of a steroid ligand, inactive prior to steroid binding
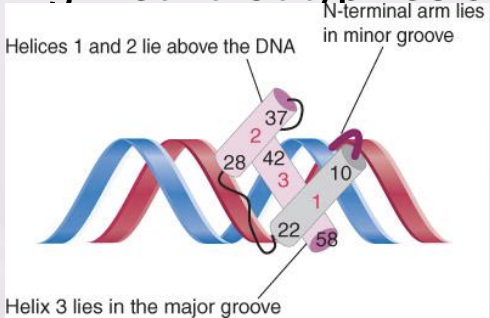
helix-turn-helix
The motif that describes an arrangement of two α-helices that form a site that binds to DNA, one fitting into the major groove of DNA and the other lying across it
homeodomain
A DNA-binding motif that typifies a class of transcription factors
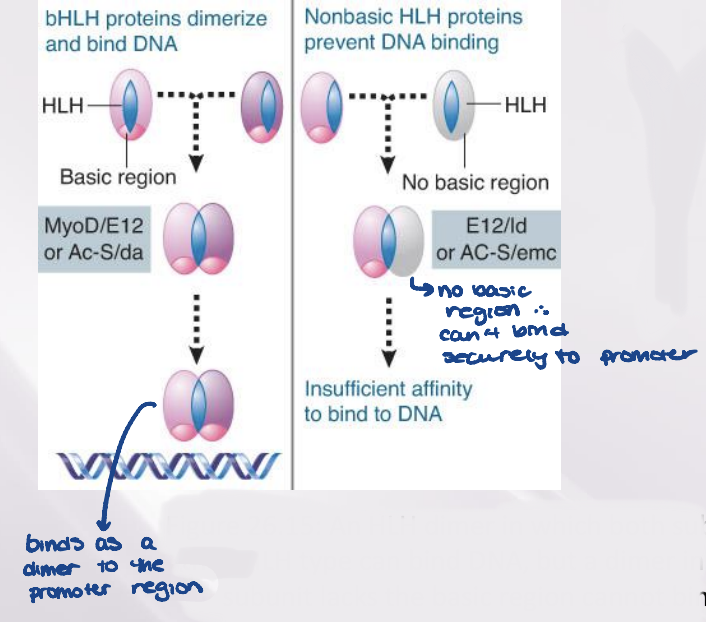
helix-loop-helix
The motif that is responsible for dimerization of a class of transcription factors called HLH proteins
A bHLH protein has a basic DNA- binding sequence close to the dimerization motif
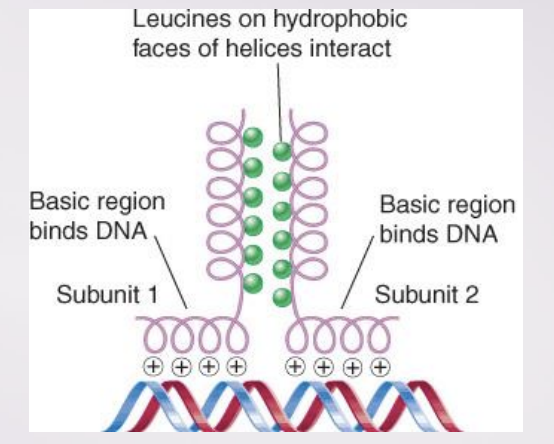
leucine zipper
A dimerization motif that is found in a class of transcription factors
bZIP (“basic zipper”)
A bZIP protein has a basic DNA-binding region adjacent to a leucine zipper dimerization motif
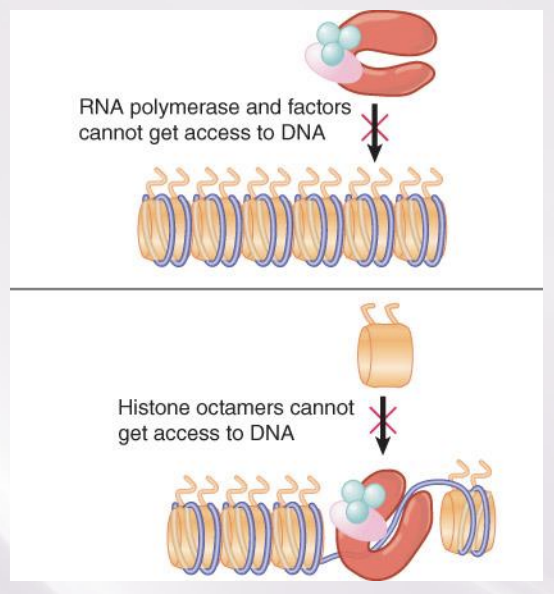
Chromatin Remodeling
The energy-dependent displacement or reorganization of nucleosomes that occurs in conjunction with activation of genes for transcription
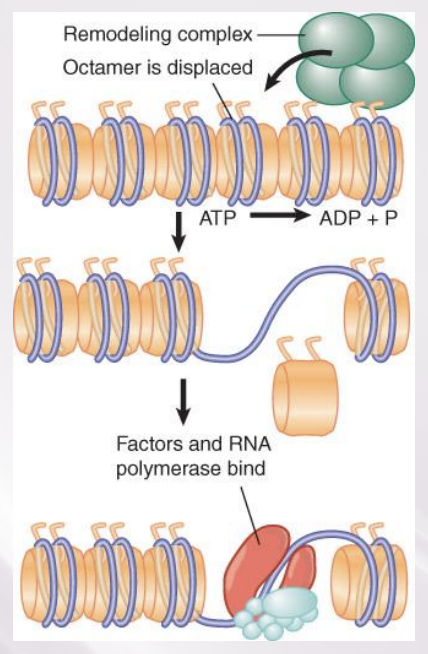
ATP-dependent chromatin remodeling complexes
Numerous ATP-dependent chromatin remodeling complexes use energy provided by hydrolysis of ATP.
what do remodelling complexes do
All remodeling complexes contain a related ATPase catalytic subunit, and are grouped into subfamilies containing more closely related ATPase subunits
Remodeling complexes can alter, slide, or displace nucleosomes
some can exchange one histone for another in nucleosome
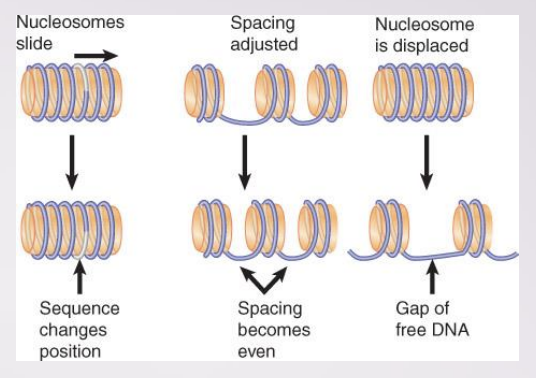
describe how Nucleosome Organization or Content Can Be Changed at the promoter
A remodeling complex does not itself have specificity for any particular target site, but must be recruited by a component of the transcription apparatus.
Remodeling complexes are recruited to promoters by sequence-specific activators.
The factor may be released once the remodeling complex has bound.
Transcription activation often involves nucleosome displacement at the promoter.
Promoters contain nucleosome-free regions flanked by nucleosomes containing the H2A variant (involved in keeping things turned off) H2AZ (Htz1 in yeast)
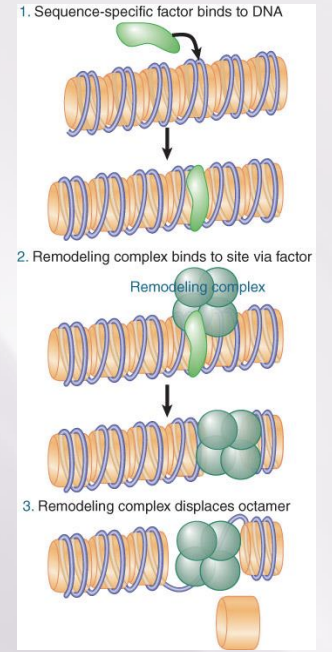
Nucleosome Organization or Content Can Be Changed at the Promoter cont.
Transcription activation often involves nucleosome displacement at the promoter.
Promoters contain nucleosome-free regions flanked by nucleosomes containing the H2A variant (involved in keeping things turned off) H2AZ (Htz1 in yeast)
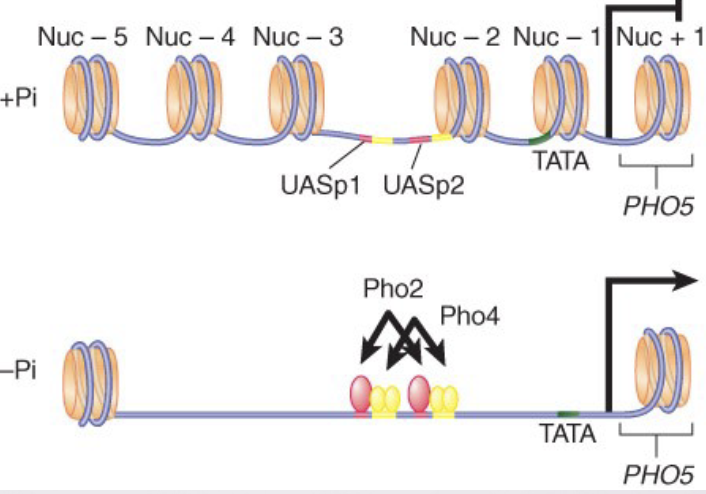
what does Histone Acetylation do?
Transcription activators are associated with histone acetylase activities in large complexes.
histone acetyltransferase (HAT) – An enzyme (typically present in large complexes) that acetylates lysine residues in histones (or other proteins)
Also known as lysine (K) acetyltransferase (KAT).
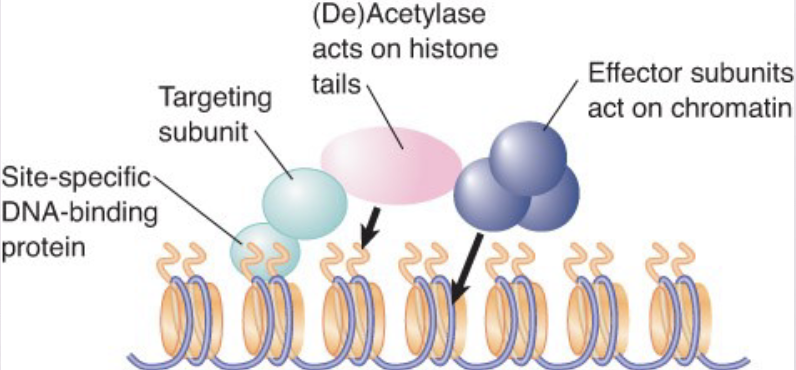
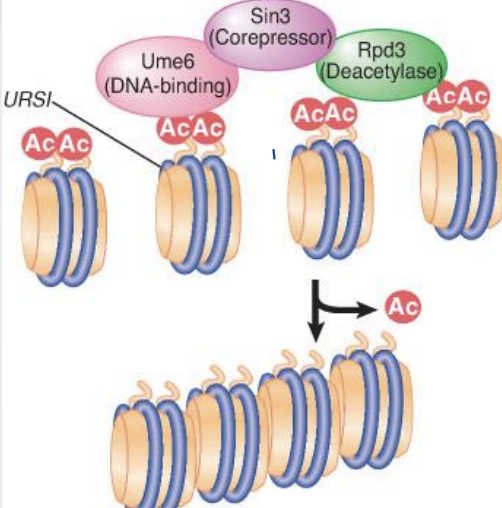
what does Histone de-Acetylation do?
Deacetylation is associated with repression of gene activity
histone deacetylase (HDAC) – Enzyme that removes acetyl groups from histones; may be associated with repressors of transcription
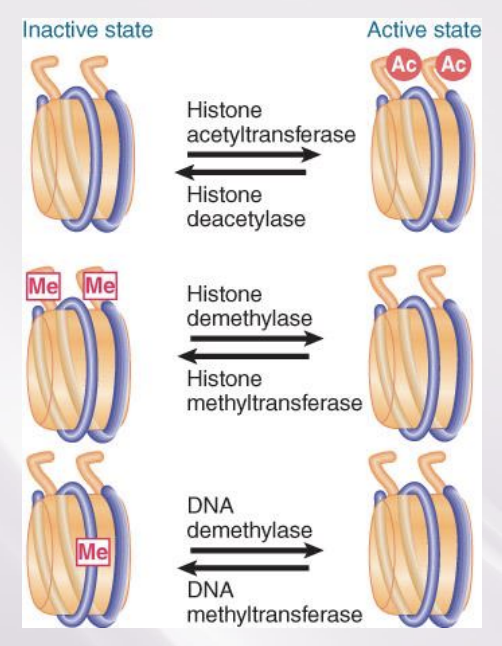
Methylation of Histones and DNA Is Connected
Methylation of both DNA and specific sites on histones is a feature of inactive chromatin.
The SET domain is part of the catalytic site of protein methyltransferases.
The two types of methylation event are connected.
Promoter Activation Involves Multiple Changes to Chromatin
Remodeling complexes can facilitate binding of acetyltransferase complexes, and vice versa.
Histone methylation can also recruit chromatin- modifying complexes.
Different modifications and complexes facilitate transcription elongation
highly methylated DNA = DNA that has been silenced for good
done once gene is no longer needed ever again
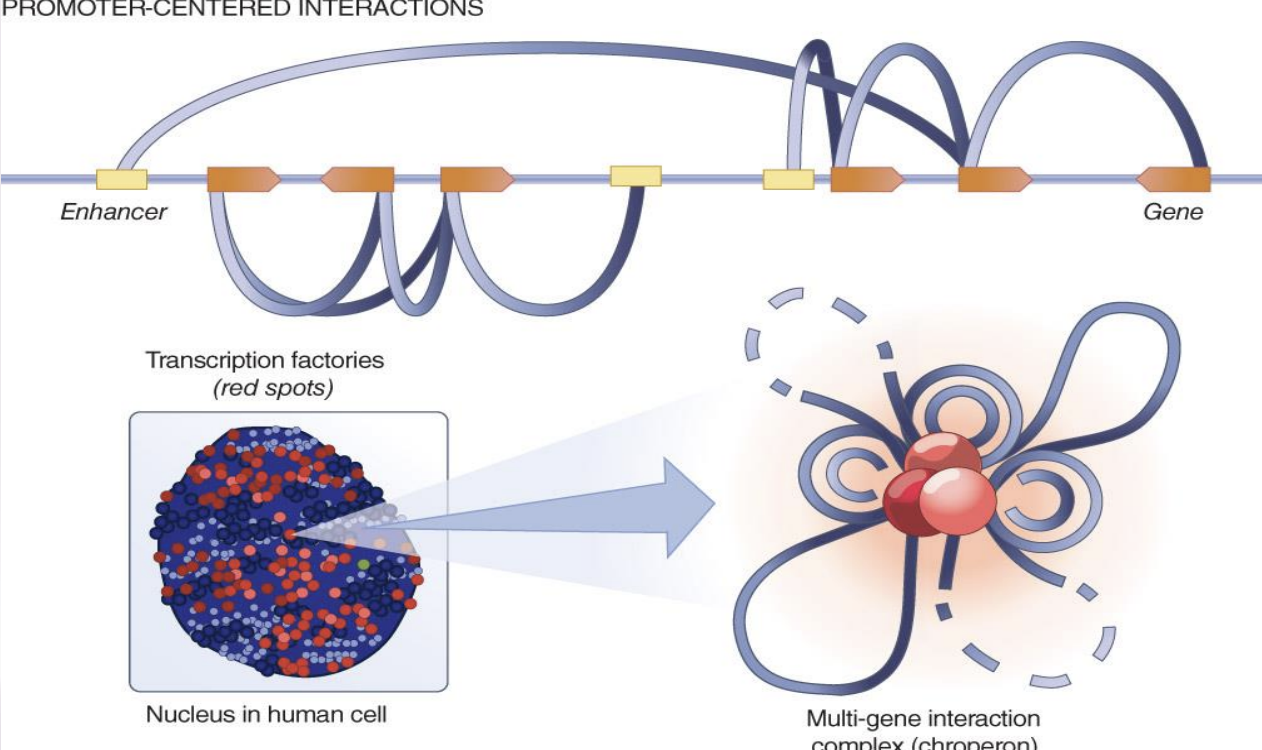
Synergistic Chromatin Interactions in Transcription Regulation
Higher-order chromatin interactions synergistically promote transcription of clustered genes. These interactions indicate a topological, combinatorial mechanism of transcription regulation