BIOL 3010 Exam 3 Flashcards
1/111
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
112 Terms
Double Strand Break
both homologs are broken at a particular point
can be caused by…
error in mitosis/meiosis
ionizing radiation
transposable element insertion/mobilization
Chromosomal Rearrangment
illegitimate recombination of chromosomes following a double-strand break
leads to mutation if not repaired
especially common in places with repeated sequences
can occur in somatic or germline cells
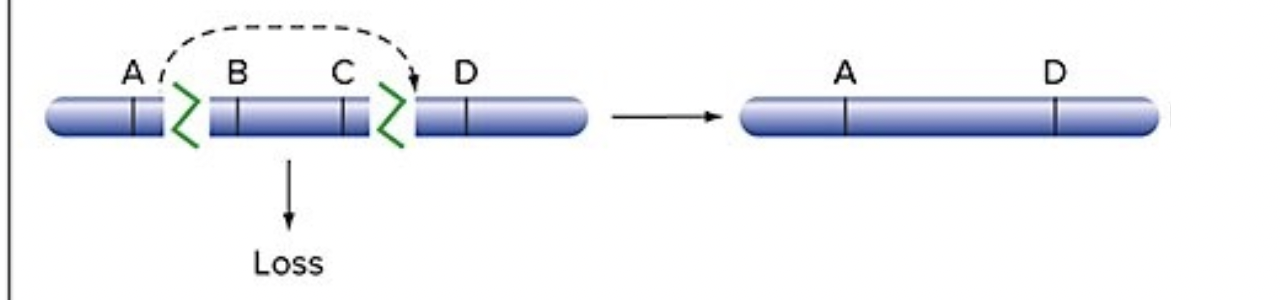
Deletion
genes in the middle of a chromosome are lost
genes on the end end up too close together
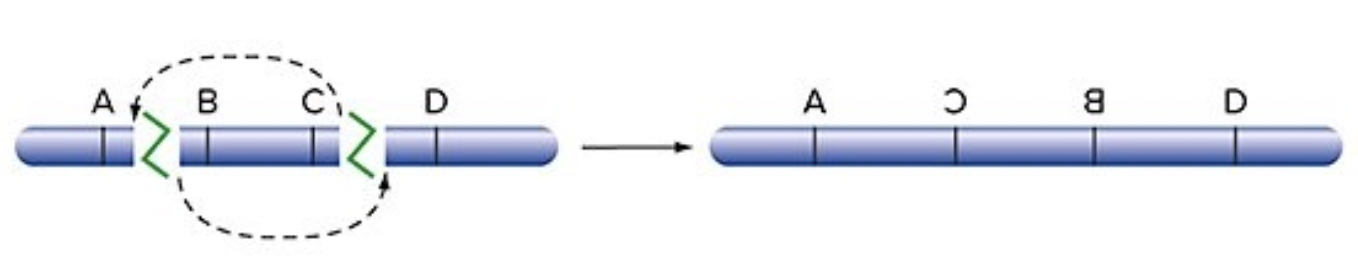
Inversion
after DSB, the middle of the gene is inserted backwards
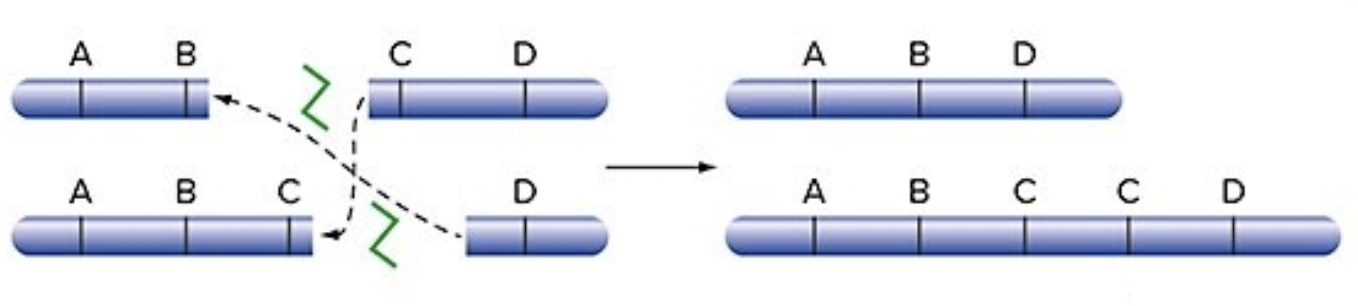
Deletion & Duplication between sister chromatids
both chromosomes are broken and joined together incorrectly
results in 1 too short of a strand (deletion)
and 1 too long of a strand (duplication
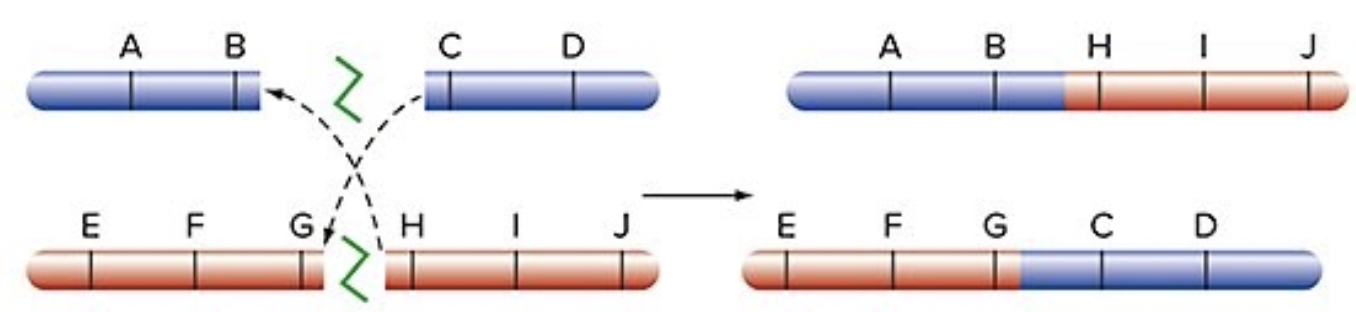
Translocation between non-homologous chromosomes
fusing the ends of incorrect chromosomes
2 completely novel genes
Acute Radiation Syndrome
-an issue in Chornobyl from the penetrating gamma rays
death by loss of proliferative stem cell population
131I exposure lead to increased risk of papillary thyroid carcinoma - why?
thyroid readily uptakes iodine - caused DSB and mutation
Outline outcome of RET/ELE1 Rearrangement
~35% of papillary thyroid cancer cases
inversion in chromosome 10 caused RET (receptor tyrosine kinase) and ELE1 (a transcription factor) to be too close together
RET no longer needed a ligand for signaling and could proliferate
What are the two ways translocations can underlie cancer?
ones affecting regulatory elements and gene expression
enhance becomes closer to a coding sequence and changes the expression
ones affecting open reading frames and protein function
different, novel combos of the genes - fusion
Homologous recombination
ideal way of DSB repair- much like crossing over process
exonuclease chews away strand around the break and the sister chromatid strand invades
replication of DNA with sister as template
almost perfect
Single Strand annealing (SSA)
less ideal way of DNA repair - at long homologies
uses a similar-ish sequence of the same strand as a template to repair the DNA after exonuclease comes in
can result in large deletions
DNA ligase fuses the backbone back together
Alternative end-joining (altEJ)
ess ideal way of DNA repair - at short homologies
allows reattachment of chromosome ends
mutagenesis rearrangement that can cause insertions and deletions
Non-homologous end-joining
pretty ideal?
joins the blunt ends of broken DNA back together
however, if there are multiple DSB there is a potential for translocation
What is special about tardigrades?
extremely resistant to ionizing radiation due to presence of damage suppressor protein
Dsup
expressed in all tardigrade cells - a protein that binds to nucleosomes to protect against damage from ionizing radiation
include single/double strand breaks and free radicals
in study testing function: cells treated with Dsup had less “oozing comet tails” than those that were not - less damage
Deficiency allele
a large deletion that results in removal of entire gene
ETV6-NTRK-3
inversion of these genes accounted for ~14% cases of papillary thyroid carcinoma
due to inversion of TF and kinase
Regions of synteny
shared gene order between or within species
can be uncovered by comparative genomics
big blocks of genes remaining together across evolutionary time
gar/chicken>car/human>human/mouse
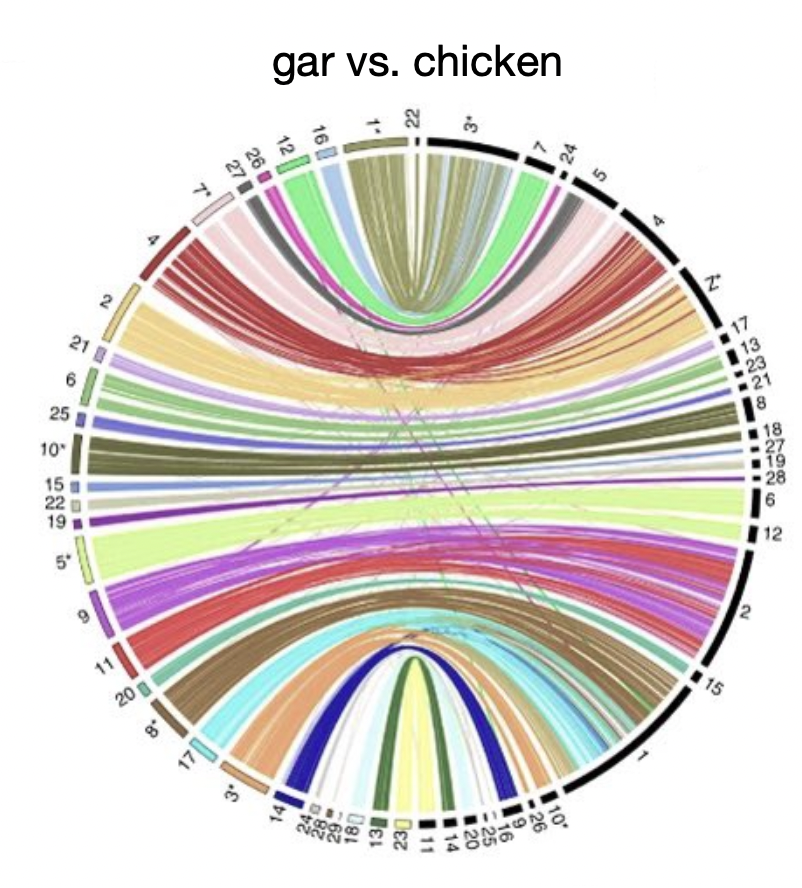
Significance of Gar?
share a lot of gene order with tetrapods, especially the chicken
it is because it has also underwent 2 WGD like tetrapods
genome is very comparable
Muntjak example takeaway
reevesi and muntjak share a common ancestor with many small chromosomes, but a fusion event in both resulted in different numbers of chromosomes in each
still very genetically similar
Muntjak- 4 large chromosomes
reevesi - lots of small ones
Paracentric v pericentric inversion
paracentric-centromere is not in the inverted portion of chromosome
pericentric- centromere is in the inverted portion of the chromosome
*we talked about pericentric
acentric & dicentric strand
acentric- after crossing over, strand without centromere
lost
dicentric- after crossing over, a strand with two centromeres
pulled in 2 opposite direction and breaks into 2- inviable
Immediate meiotic consequence of inversion:
heterozygote for inversion has reduced fertility
Long-term meiotic consequence of inversion:
there are no crossings over at sites of inversion
genes not broken up so they can acquire new mutations together
Super gene
alleles always stay together and do not participate in crossing over- result of inversion
ex. Ruff bird
Ruff bird example significance
males have 3 phenotypes: wildtype, satellite, and Faeder
used PCR priming to discover that satellite and faeder are heterozygous for an inversion
has supergene that has acquired multiple mutations such as ones in melanocortin 1 receptor required for pigment
Inversion loop
when a non-sister chromatid has an inverted segment, it must take on an inversion loop during crossing over to line up the base sequence
Homologous gene
genes that have shared ancestry
orthologues
homologous genes of different species
ex. between gar and chicken
paralogues
homologous genes in same species due to a duplication event
ex. Hox genes
Copy number variants
aka CNVs
major class of genome structural variation
we all have different amount of copies of these regions
deletions and insertions associated with disease
Rhodesian ridgeback dog example
“ridgeback” trait is dominant and predisposes dog to dermoid sinus
there is a duplicated region that has 3 Fibroblast Growth Factor genes which play role in hair development
results in reversal of hair direction
Fibroblast Growth Factor
3 extra copies in Rhodesian ridgeback mutant
secretes growth factor that results in reversal of hair direction if you have too many copies
Dermoid Sinus
congenital disease that is associated with ridgeback phenotype of Rhodesian dog
Whole Genome Duplication
occurs through different abbreviations of the cell cycle
polyploid cells are common and preferred in some cases
has happened throughout evolutionary history
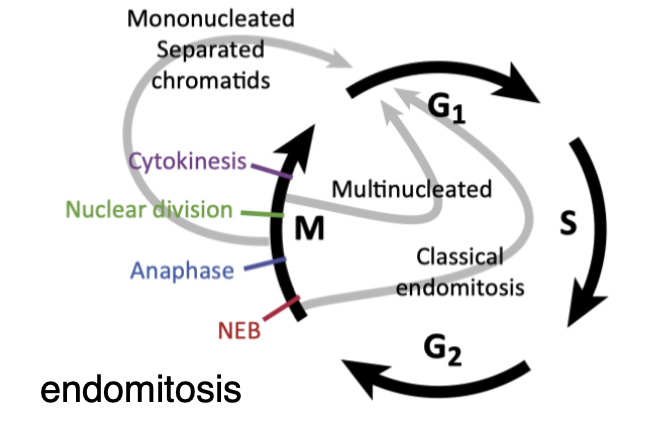
Endomitosis
mitosis occurs but doesn’t result in cellular division
if the nucleus divides- multiple nuclei
if not - 1 nucleus
Endocycle
cell cycles between the G1 phase and S phase repeatedly without mitosis
Rediploidization
after a whole genome duplication, some of the duplicated genes are maintained, but most are lost
because of redundancy in function
if no selection to maintain both copies they are lost
ex. zebrafish underwent an additional duplication, but only has 30% more protein-coding genes than humans
WGD’s in vertebrate lineage
2 WGD in tetrapods and Gar
3 WGD in teleosts, some underwent a 4th
WGDs in Hox genes
conserved synteny with evidence of WGD in hox genes
found as 1+ clusters of multiple genes
remain synteny within/between species even though there were losses and duplications
ohnologues
homologous genes arising by whole genome duplication
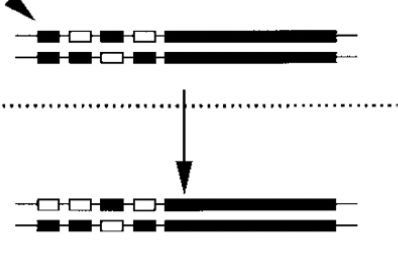
Neofunctionalization
when cis regulatory element involved in duplication..
with additional mutation, results in complete loss of new gene
nonfunctionalization
copies of alleles diverge functionally because one of them acquires a new function domain
made possible because the top gene is covering original function
additional mutation is in regulatory element
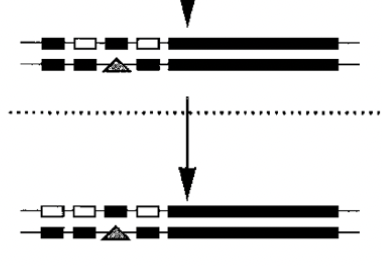
subfunctionalization
loss of an additional regulatory element
to fulfill the initial function, you now need two different genes
Why do polymerase errors lead to expansion/contraction?
especially in repeated sequences, it is easy for DNA polymerase to slip forwards or backward
-repeats aren’t paired correctly and a portion of nt sticks out
-one round of replication happens: have 1 larger and 1 shorter copy
-results in the divergence of alleles
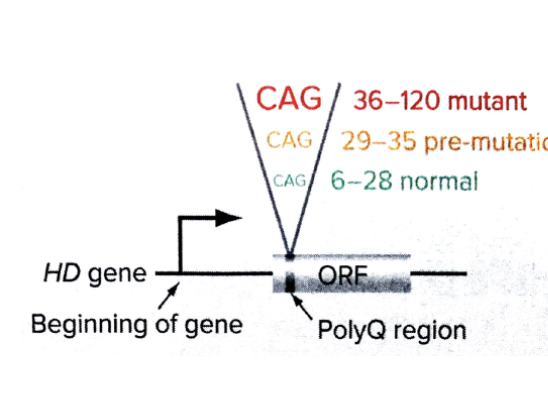
Huntington Disease
poly Q expansion
symptoms
late onset neurogenerative disorder typically appearing ~30/40
progressive chorea, defects in speaking and swallowing, mental decline
because of a glutamine expansion in exon 1 of the huntington protein
Point mutations
single nucleotide additions, deletion, or substitution
can result from polymerase errors
Transversion
point mutation going prom purine←> pyrimides
ex. A to T or C, T to G or A, C to G or A, G to T or C
Transition
point mutation between purines or pyrimides
ex. A - C, G - T
Replication-coupled DNA repair
DNA polymerase has exonuclease activity that proofreads sequence 3’ to 5’
corrects vast majority of point mutations
takes out wrong base, allows for new, puts correct one in its place
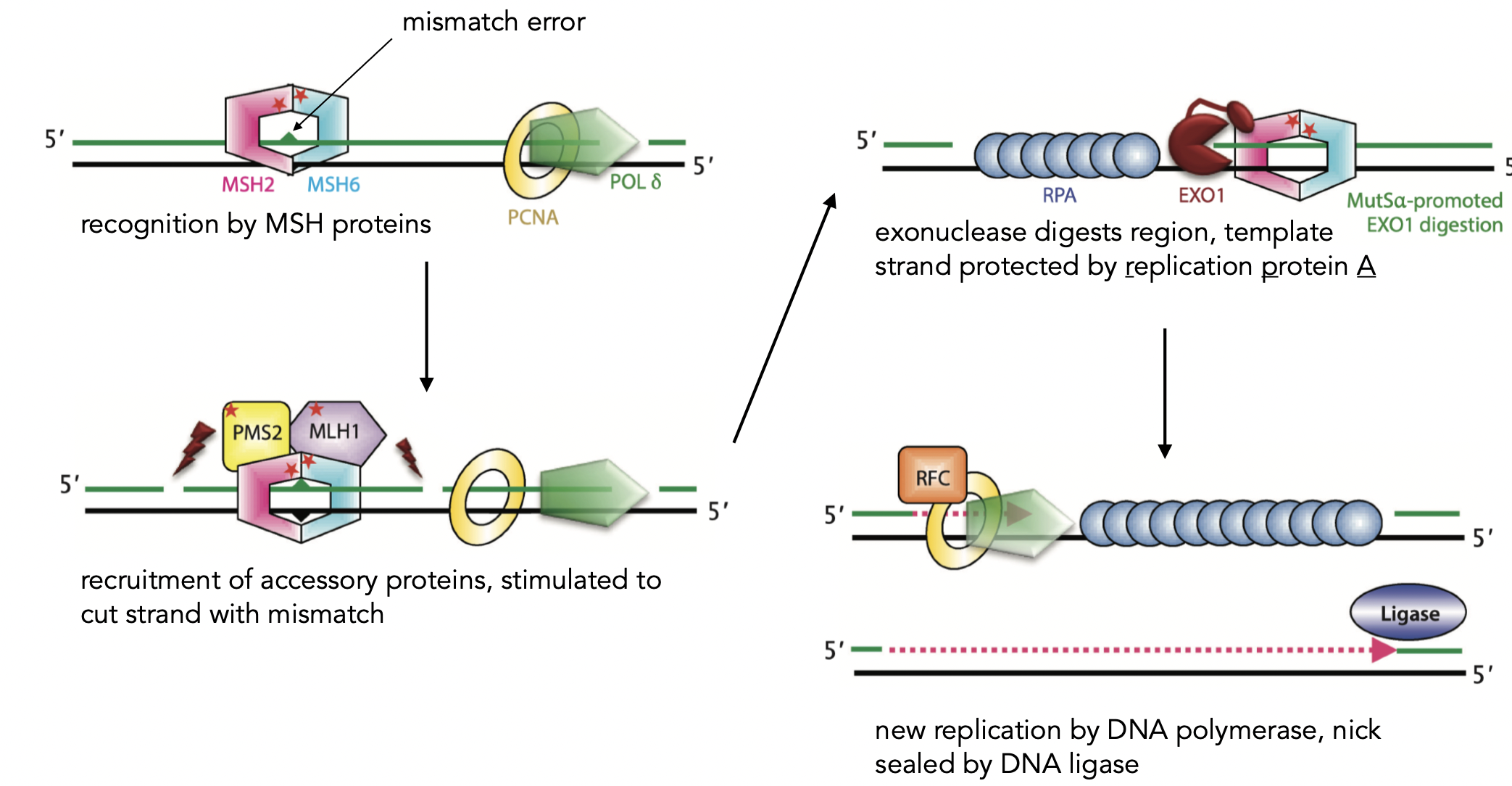
Mismatch repair Pathways
corrects polymerase errors and DNA damage
associates with DNA and scans for missmatches
exonuclease digests region→ template strand protected by RPA → recruits accessory proteins that are stimulated to cut strands → new replication by DNA polymerase → nick sealed by DNA ligase
DNA Damage: Hydorlysis
depurination - guanine released
deamination - the amino group is taken off cytosine and it turns into uracil
after replication, turns site into TA
DNA Damage: Oxidation
oxidized guanine looks different to polymerase - pairs with A
after replication, results in TA site
Nucleotide/base excision repair
general steps:
proteins scan for problems
portion chewed away by exonuclease
polymerase lays down correct nucleotides
backbone ligated together
Crosslinked DNA (intra v inter)
inappropriate covalent bonds within (intra) or between (inter) strands of DNA
blocks transcription and replication
intrastrand fixed by base excision repair
interstrand requires a big protein complex to fix it during replication
Fancomi anemia
symptoms:
growth retardation, hyperpigmentation, renal/skeletal abnormalities, mental impairment, hear defects, cancer risk
Why?
due to damage resulting from interstrand crosslinked DNA
de novo mutations
mutations that arise in the germline of the parent
observed at a 4:1 ratio in paternal gametes
can be calculated through a trio sequencing test
trio-sequencing
whole genome sequencing of the mom, dad, and progeny to determine if a mutation arose in the germline or somatic cells of the parents
found that mutations 4x as likely to cone from paternal allele and increased as paternal age increased
Why are de novo mutations more common in paternal gametes and aneuploidy mutations more common in female oocytes?
de novo is more common in males because there is continuous replication of DNA in the sperm cells
more likely to have error
aneuploidy more likely in female oocytes because of delayed completion of meiosis
Loss-of-function alleles: null
mutation resulting in complete loss of gene product
Loss-of-function alleles: hypomorphic
mutation causing gene to make less product or be less active than wildtype
Gain-of-function alleles: overexpression
mutation causing higher expression of a gene than the wildtype
Gain-of-function alleles: hyperactivity
mutation causing higher activity than wild-type in the product itself
ex. a receptor no longer needs a ligand to produce product
Gain-of-function alleles: neomorphic
mutation causing a gene to take on a new expression domain/protein activity
Advantages of drosophila as model organism
short generation time
many progeny
can evaluate exoskeleton under a microscope
only 4 chromosomes - often polytene
many similarities in genes/functions with vertebrates
Polytene chromosome
natural polyploid where multiple copies of the same gene are present in the same area
advantageous for examination of model organism
Disadvantages of drosophila as model organism
stocks must be maintained as “living” cultures
N-ethyl-N-nitrosourea (ENU)
an alkylating, mutagenic agent that increases mutation rate per locus past what would be spontaneous
to use mutation as a genetic tool
adds ethyl group to induce random base changes throughout the genome
Ethyl methane sulfonate (EMS)
an alkylating, mutagenic agent that increases mutation rate per locus past what would be spontaneous
to use mutation as a genetic tool
adds ethyl group to induce random base changes throughout the genome
Main goal of Nusslein-Volhard & Wieschaus
sought to identify genes required for segmentation, anterior/posterior or dorsal/ventral differences of Drosophila
conducted forward genetic screens to identify certain genes important for embryonic development and body patterning
Forward Genetic Screen
mutants identified by phenotypes of interest and then genes responsible for phenotypes are determined later
ex. drosophila experiment
Reverse Genetic Screen
targets a specific gene and see if it produces specific phenotype
Saturation of Genetic Screen
when you are finding fewer mutation at new genes
saturated when new alleles are at loci for which mutants have already been found
could get new alleles, but on the same gene
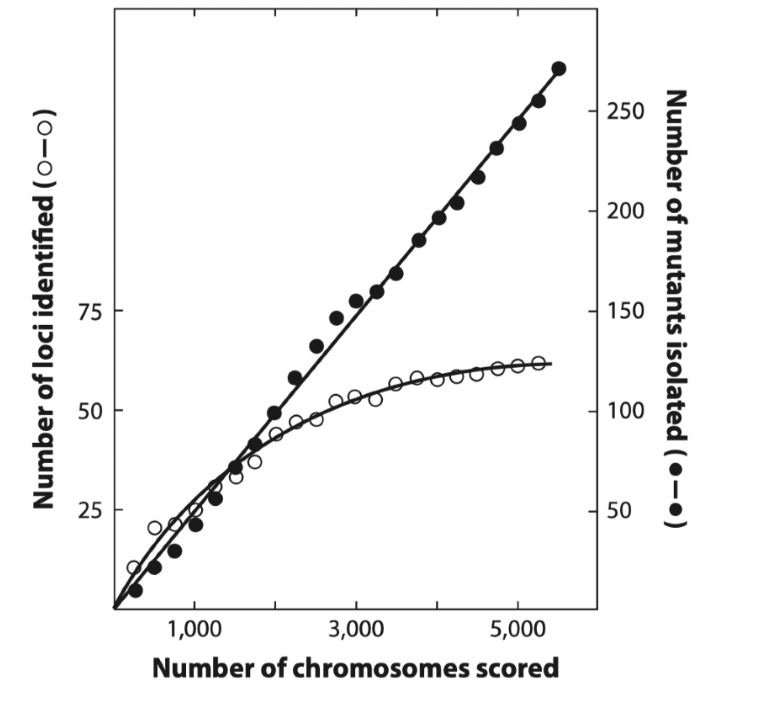
Gap gene mutants in Drosophila
mutants have large regions of banding missing
ex. krupel, knirps
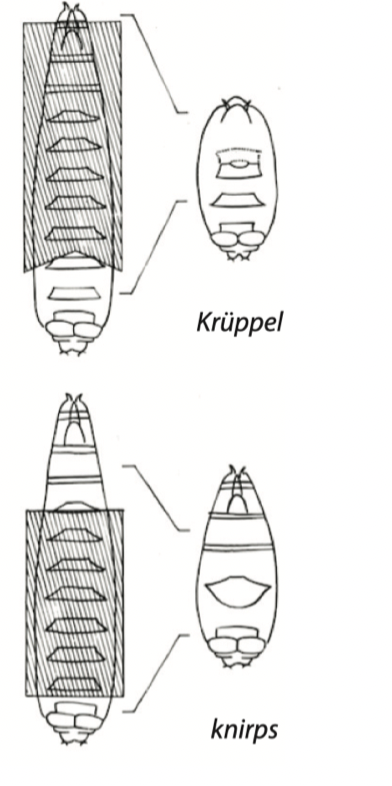
Pair-rule gene mutants in Drosophila
mutants have alternating segments lost, the defects are in pairs
ex. even/odd-skipped, paired, runt
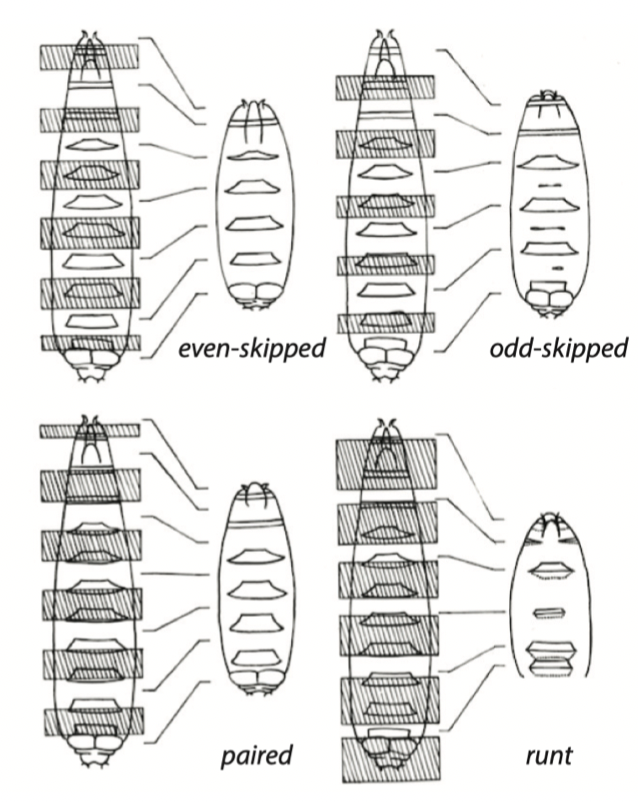
Segment polarity mutants in Drosophila
portion of the segment is missing or altered
ex. gooseberry, patched
General cascade of genes impacting body development in drosophila
hierarchy of genes that establish body plan
cytoplasmic polarity → hunchback protein gradient → gap genes→ pair-rule gene → segment polarity genes
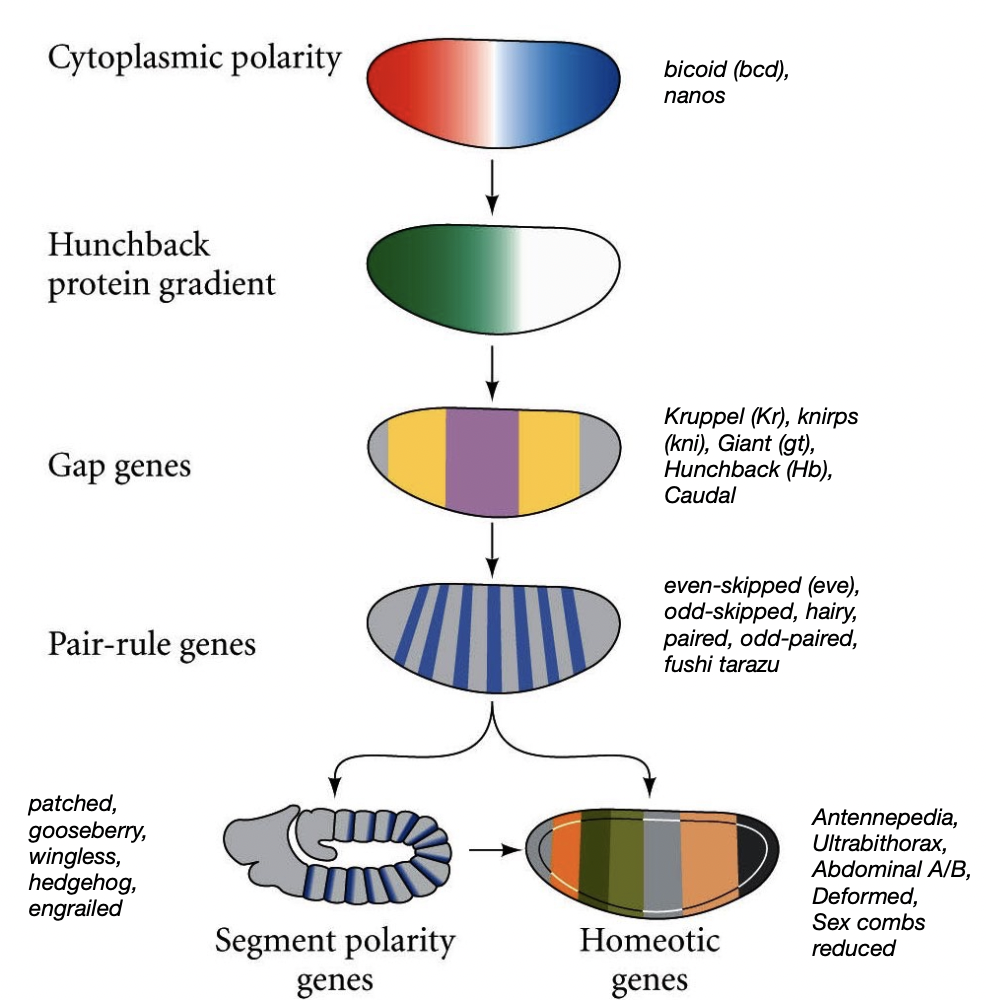
Outcrossing
crossing a heterozygote F1 with a homozygous parent
Complementation Test
use to see if two mutations are alleles of the same or of different genes
cross two mutants and get a mutant progeny
the same gene!
cross two mutants and get wild-type
different gene! heterozygous
only works for recessive alleles
Meiotic mapping
uses recombination that occurs during meiosis to narrow down the location of a mutation in the genome
recombination frequency and cM
recombination frequency = # of recombinants due to crossing over/ total alleles
multiply this by 100 to calculate the genetic distance in cM
follows idea that the closer two genes are on a chromosome, the less likely they are to be separated during crossing over
what does little crossing over between two points indicate
genes are really close together
Loci markers
Identifiable physical location on the DNA where the sequence is variable and can be detected
provide a landmark on a gene that we can compare mutant gene to by seeing which alleles at marker loci across genome phenotype most closely associates with
What are stretches of DNA that can be used as marker loci?
1) microsatellites
repeated stretched of CA can be variable across individuals
can detect size differences on gel
2 bands = heterozygote 1= homozygote
2)Restriction Fragments
at a locus, specific site is cut by particular restriction enzyme
homozoygous if 1 or 2 bands, heterozygote if 3 bands
3) Use of PCR and Sanger Sequencing
shorter wave at a site indicates variable alleles
in general we want to look for nucleotides that are variable within population
Effect of gene redundancy and compensation
some mutant genes won’t show phenotype
other genes may be picking up the slack
fusion
Smaller chromosomes join together to make larger ones
fission
Larger chromosomes split to becomes smaller ones
Gray
a measure of radiation
genotyping
process of determining differences in the genetic make-up of an individual by examining the individual's DNA sequence using biological assays and comparing it to another individual's sequence or a reference sequence
important in meiotic mapping
ligase activity
Joins DNA strands together by catalyzing phosphodiester backbone to form a bond
important in DNA repair!
spontaneous mutant
mutations are randomly identified by a breeder, no mutagenic agents were intentionally used to cause damage
very rare and hard to predict
informative meiosis
During genetic mapping, when an individual is heterozygous for an allele, their gametes will undergo crossing over to create two distinct sites where genetic backgrounds are switched
You can use these to determine relative position of gene on chromosome
inbreeding heterozygote - 2
outcrossing w homozygous parent - 1
recombinant
Chromosomes that carry a mix of alleles due to crossing over of non-sister chromatids
Cans use the frequency to calculate cM, genetic distance
complementation
When mutations are in different genes - outcome is a heterozygous wildtype progeny when crossed
non-complementation
When mutations are on the same gene, the result when crossed is a mutant
allelism
The same gene having variable sites
important for marker loci to have variability so you can compare genetic backgrounds
phenotypic rescue
An important strategy for identifying the right gene of interest
By either replacing sequence itself or adding protein activity
Switching the mutant phenotype to the wild-type by adding in what gene is lacking
revertant allele
A wild-type allele present that was once a mutation
Researcher targeted the gene to alter sequence and see if the issue was resolved
transgene
cloned segment of DNA in a test tube that researchers have inserted into the genome of an organism
Could be used to produce lacking protein activity/products
genetic background
all of the alleles of genes in an organism’s genome; the set of unknown modifier genes that influence the action of the known genes that control specific aspects of phenotype
needs to be different between mutant and marker loci so you can compare to see where crossing over took place