EVE 100 Midterm 2
1/83
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
84 Terms
Dominance
describes the effect of an allele on a phenotypic character when it is paired with another allele in a heterozygote condition —> higher fitness
takes longer to reach fixation since the recessive alleles hide in the background
Advantageous dominant alleles increase in frequency more rapidly since it is expressed in both homozygous and heterozygous genotypes
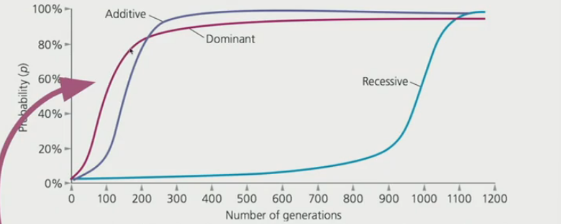
Recessive alleles
Hide behind the dominant allele even when advantageous
advantageous recessive alleles take a long time to increase in frequency since when at lowe frequencies, it is present in the heterozygous form → invisible to selection
Advantageous recessive alleles can reach fixation since they hide in the background
After the advantageous dominant allele attains high frequency, the alternative disadvantageous recessive allele is slowly eliminated → rare recessive alleles occur mostly in heterozygous forms -→ invisible to selection

Fitness
Measure of the “relative success of a population, genotype, and alleles
measures how often thinsg reproduces
defined by the probability of survival and the avergae number of offspring
best applied to a set of entities → all individuals in a population or individuals with a given genotype
MOST fit genotype = relative fitness of 1
LEAST fit genotype = relative fitness of 1-s (s = selection coefficient)
HETEROZYGOUS genotype = relative fitness of 1- hs (h - dominance coefficient) → determines how far the heterozygous genotype is from the homozygous phenotypes
h = 0 → A1 is dominant
h = 0.5 → A1 is additive
h = 1 → A2 would be dominant (A1 is recessive)
Finding relative fitness
absolute fitness/the highest absolute fitness —> needs to be ain a decimal form less than or equal to 1
Selection coefficient
finds how fit something is from the MOST fit organism
most fit h and s coefficients help us understand how dominance affects fitness
Decent of modification
genetic changes in populations over timeM
Mutations
Mutation rates per base pair are low —> but the number of new mutations per individual may be large
humans have a mutation rate of ~2.5 × 10^-8 mutations per generation → BUT the number of new mutations PER individual may be large
Humans have around 160 new mutations per transmission (each of us has 160 unique mutations that other people do not have)
Mutations are random but common
Mutation types:
single base pair mutations
translocations
deletions/duplications
whole genome duplications
inversions
transposable elements
retrovirus insertions
chromosome fission and futsion
Mutations can either be beneficial, deleterious or neutral —> they can also have small or large effects —> they can also be dominant or recessive
most mutations are directly selected out of a population
Distribution of fitness effects (DFE)
this looks like a continuous ditribution
most effects of mutations are small —> most new mutations reduce fitness ( a little)
fitness effects of mutations —> less than 1 = deleterious and decrease fitness and fitness greater than 1 are probably beneficial mutations
Heterozygosiry = H
this is the fraction of sites that are heterozygous → more heterozygous genotypes = higher heterozygosity
humans have a heterozygosity of 0.1 → therefore diversity within humans are low
higher heterozygosity = lower offspring (ex., hybrids survive but rarely reproduce)
3 reasons for how genetic and phenotypic polymorphism is maintained
→ polymorphism: different phenotypes = different genotypes
balancing selection: selection sometimes acts to maintainvariation
Mutation selection balance: the rate of mutation and the rate of selection are both similar emough that the mutations are not gotten rid of due to the rate of new mutations being too fast
mutation-drift balance ( neutral theory): genetic drift
Balancing seelction
when selection can maintain balanced polymophism in a population
overdominance or heterzygote advantage: Heterozygotes for an allele have a higher fitness than either homozygote
negative frequency-dependent selection: selction favors rare morphs —> states that it is better to be different → this increases trait diversity
spatially or temporally variable selection: variation in selection across space or time maintains genetic variation
Overdominance and Underdominance
Best genotype = combination of 2 different types of alleles
over mean fitness when there is an intermediate frequency of alleles → p is in the middle → this maintains polymorphism for more favorable genotypes → heterozygote is the BEST
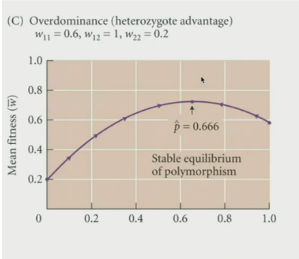
Underdominance: when the heterozygote individual is the worst → the AA or aa genotype is the best
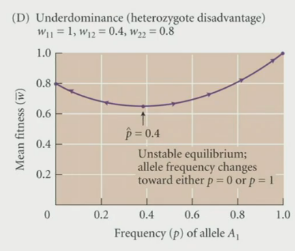
Example: sickle cell anemia → the patient is less susceptable to malaria when they have a heterozygous genotype for those with mild anemia (Ss = mild anemia) → (SS = sickle cell anemia) → thisis because the RBCs have shorter life spans → this gives the plasmodium less time to reproduce
If a heterozygote has the HIGHEST fitness = heterozygote advantage = overdominance
Negative frequency-dependent selection
genotype/phenotype has a fitness advantage when it is rare, but its fitness decreases as it becomes more common
fitness is negatively correlated with frequency → lower frequency = higher fitness
rarity is an advantage
Spatially and temporally variable selection
variation in a phenotype of genotype is maintained by variation in selection patterns across environments → one allele has a fitness advantage in one environment while another allele has a fitness advantage in another environment
e.g. Bird beak shapes → change yearly based on drought → smaller or bigger → adapt to time and environmental conditions
spatial and temporal selection can occur at the same time
local adaptation = a form of variable selection
Things that determine the amount of variation in a population
mutation selection balance
mutation and inbreeding (gene flow) between populations
Ge
Gene flow
homogenizes allele frequencies UNLESS selection prevents it → if it does then we get a migration selection balance → how gene flow (migration) and natural selection interact → equilibrium that forms when:
Migration brings alleles into a population → continuously introduces new variants to a population
Selection works against those alleles
two forces balance out so the allele stays at an intermediate frequency instead of being lost or fixed
If selection eliminates deleterious variants, variation can be maintained
Mutation selection balance
mutations continuously introduces new variants to a population → if selection eliminates the deleterious mutations → variation can be maintained
mutation = MAIN SOURCE or genetic variation
stronger selection = rate of taking away deleterious variatns in the population is faster
Weaker selection = rate of taking away deleterious variants in the population is slower
Mutation selection equilibrium
Deleterious alleles that dont have a too strong of an effect in the heterozygote can be maintained at low frequenes in the population → recessive deleterious alleles
Mutation introduces new variation into a population
Random genetic drift determines heather a neutral allele will be fixed or (usually) lost
At equilibrium, there is a balance between mutation and genetic drift
Inbreeding depression
reduces fitness of inbred individuals due to deleterious mutations
reduced fitness in offspring that results from mating between relatives.
more recessive mutations should segregate at high frequencies under mutation-selection balance
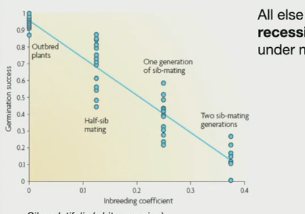
Expression of deleterious recessive alleles
Normally, recessive harmful mutations are “hidden” in heterozygotes.
Inbreeding makes offspring more homozygous, so these bad alleles get expressed.
Example: genetic disorders in royal families with lots of cousin marriages.
Loss of heterozygote advantage (overdominance)
Some genotypes (heterozygotes) are fitter than either homozygote
nbreeding reduces heterozygosity, lowering this benefit
The closer the parents are to each other (genetically) → the lower of the mean lifespan of children → this is due to everyone having these recessive negative deleterious mutations hiding in our genomes under our dominant alleles
Polymorphism
Overdominance
Having A2 always better than A1 → directional selection
Overdominance: heterozygous genotype is the most fit → aka heterozygous advantage
Eg. sickle cell anemia → maintains variation of an allele in a population
Mutation - selection balance → reasons we might see variation in a population
“Giant sink of all of our alleles in a population”
→ lots of allele variation = full sink
→ little allele variation = empty sink
Need to induce mutations (faucet → fills the sink up)
New alleles → everyone looks more different
Selection: can remove alleles in our population (drain) → “directional selection” that is natural selection
We can get an equilibrium of our variation → called the Mutation Selection Equilibrium
We need to know μ: rate of mutation
s = selection coefficient
h = dominance coefficient
q(eq) = mutation rate (μ)/ ((selection rate(s) * dominance coefficient (h)))
Balance between mutation → Genetic drift
we dont believe in selection → we believe in a more random process where we take variance outside of the population
instead of selection we get drift
mutation drift equilibrium
Ne = (N = number of things we see → E = means effective)
Ne = effective population size → if mutations add things to a small population → “size of an idealized population (one that follows Hardy–Weinberg assumptions perfectly) that would experience the same amount of genetic drift, inbreeding, or loss of diversity as the real population”
larger population size = more even probability of allele frequency
genetic drift → more effective in smaller populations
neutral alleles are alleles that don’t impacts fitness → synonymous mutations and codons → change in third codon doesn’t change the fitness of an organism
selection
Neutral theory of molecular evolution
most mutations are deleterious and are lost immediately
Most observed molecular polymorphism and genetic substitutions are neutral → consistent with high levels of genetic polymorphism and molecular clock
Frequency of neutral alleles are controlled by:
Mutation rate
Genetic rate
Mutation - selection balance
occurs when new mutations introduce deleterious alleles at the same rate that natural selection removes them
new mutations create new alleles
selection acts to remove deleterious alleles
Definition: Frequency of deleterious alleles is controlled by:
mutation rate (μ or aka u)
selection coefficient (s) and dominace coefficient (h)
qeq (equilibrium frequency) = u/(sh)
Mutation-drift balace (neutral theory)
most mutations are deleterious and are lost immediately
Most of the observed molecular polymorphisms (like enzyme amino acid sequence variation) and genetic substitutions are neutral
claimed to be consistent with high levels of genetic polymorphism and the molecular clock
Mutation drift quilibrium (MDE)
Mutation introduces new variation into a population
random genetic drift determines whether a neutral allele will be finzed or (usually) lost
at equilibrium, there is a balance between mutation and genetic drift
mew meutral alleles arise continually by mytation → many are usually lost by genetic drift while others drift to higher frequencies and persist for some time in a polymorphic state before they are lost or fixed
→ definition: under the MDE, the amount of variation introduced by mutation = the amount of variation lost due to drift → this is dependent on:
Ne = effective population size
Myu = mutation rate to new neutral alleles
at equilibrium under MDE → heterozygosity = H = 4Ne*(myu)
→ in larger populations:
the rate of genetic drift is slower
The input of new mutations is higher
the level of neutral polymorphism is higher
Hardy-Weinberg assumptions
diploid secual population with dicrete generations
allele frequencies equal in both sexes
mendelian segreghation of alleles
random mating
no natural selection
no mutation
no migration
large population sizes → no drift
Genetic drift ( and selection )
These are the two most important forces affecting allele frequency changes in a population
genetic drift: fluctuations in allele frequencies that occur by chance → particularly in small populations as a result of sampling error
It can also change the frequency of neutral alleles (alleles with no effect on fitness)
can change the frequency of alleles under selection only if selection is WEAK
Changes by genetic drift are random in direction
results from a random sampling error → can be thought of as randomly sampling gametes to form the next generation → allele frequencies increase or decrease by chance
rate of genetic drift is higher in smaller population → in large populations they drift slower
droft leads to a loss of heterozygosity as it causes loss or fixation
drift is higher when the variance for reproductive success is higher
Neutral allele
an allele with no effect on fitness compared to other alleles at that locus
only 2% of our genome encodes for proteins
changes outside of exons (coding regions) can be completely neutral if they do not disript gene regulation
examples
synonymous change in a codon → the a.a doesnt change since the third base in a codon are usually neutral → the are more changed by drift than selection
non-synonymous change → replaces one amino acid with a functionally similar one → produces a large change in a phenotype oin which selection no longer acts → can cause pseudogenes
Non-coding regions that make up most of the genome
RNA genes
pseudogenes
mobile genetic elements
repetative DNA
Population bottlenecks
thisis when a popualtion size is dramatically reduces
can be caused by a widespread outbreak or diesease, high hunting pressure, extreme natural evcents, habitat shifts
this can be a short-term reduction in population size with long-term effects on allele frequencies → rare alleles are likely to be lost
some rare alleles can by chance, drift to high frequency within the population
lowers heterozygosity
this is a form of inbreeding
Inbreeding
two alleles are identical by decent (IBD) if they are the same alleles due to inheritance from a very recent shared common ancestor
inbreeding coefficient (F): thisis the probability an individual inherits two alleles identical by descent at a locus
F = 1 → completely inbred
F = 0 → completely unrelated
F = 0.25 → siblings
F = 0.125 → ½ siblings
F = 0.0625 → first cousins
F = 0.0156 → second cousins
F = proportion of the genome that is homozygous due to inbreeding
consequence of inbreeding:
The frequency of homozygotes is higher
The frequency of heterozygotes is lower
Effective population size
Ne = effective population size = the number of individuals in an idealized population that would produce the same amount of genetic drift as in the real (census) population
Ne«N (census population size)
The rate of genetic drift/ loss of heterozygosity is increases if:
The population often crashes to a small size
there is high variance in reproductive success among individuals
Factors effecting population size
variation in the number of progeny among males, females, or both
sex ratio away from 1:1
Natural selection (variance in reproductive success)
inbreeding
fluctuations in population size
Probability of fixation for a new allele
a new allele has just arisen by mutation that has a frequency (p) of: 1/2N
This is the probabilty that the new allele reaches fixation
fixation is when the alle frequency p = 1
loss is when the allele frequency p = 0
Larger populations = longer time to reach fixation
Substitution rate per generation
substitution rate per generation = number of new mutations per generations * the chance of a mutation fixing
→ 2N*(myu) *(1/2N) = myu per generations → we can eliminate N → therefore mutation is independent of population size
Levels of contraint
neutral theory claims that most new mutations are deleterious and are lost immediately → only neutral mutations contribute to substitution
neutral mutation rate → myu - myu(t)(1-C)
consistent with neutral theory → slower rate of substitution at more constrained sites
contraint (C): the proportion of mutations that are too deleterious to contribute to polymorphism or substitution
therefore: 1-C = the proportion of mutations that are neutral → in regiouns like pseudogenes → we expect the contraint to be low (1-C = ~1)
dN = non-synonymous substitution/site = C
dS = synonymous substitution/site = 1-C
EXPECTED dN/dS for a typical gene = less thjan 1
A SLOWER rate of substitution at more CONTRAINED sites is seen
effective population size will largely determine the levels of contraint
Nearly neutral alleles
chance (genetic drift) plays a role in the fate of all allleles but matters more for neutral or very near to neutral alleles

alleles that aren’t strictly neutral, but their effect on fitness is very small, so whether they behave like neutral alleles depends on population size
Selection is weak: These alleles have tiny positive or negative fitness effects.
Drift matters: Random genetic drift can override weak selection in small populations.
Implication for molecular evolution:
Some slightly deleterious alleles can fix in small populations, contributing to molecular change.
Explains why the molecular clock isn’t perfectly constant across all species.
The dN/dS ratio
most genes will be evolving slowly since they are under contraint
expected dNdS for a gene encoding an uncontrainted protein = 1 (number of non-synonymous = number of synonymous substitutions)
genes under consistent directional selection have a dNdS GREATER than 1 → this is rare → this is positive selection for a gene
most non-synonymous mutations are deleterious and removed by negative selection
larger Ne = selction is more effective at removing “nearly neutral” alleles ( weakly deleterious alleles) so the dN/dS ratio would be smaller
only calculated for CODING REGIONS → need to use a condon chart to determinine they are synonymous or non-synonymous
dN/dS → typically much smaller than 1
C-value paradox
c-value paradox: no direct correlation between phenotypic complexity of eukaryotic organisms and the size of the genome (larger genome size does not mean more complex)
There is no consistent relationship between the genome size (C-value) and the organism’s complexity
Transposable elements
this is a short DNA sequence that makes copies of themselves (they act like DNA parasits -→ AKA selfish DNA)
(a and b) DNA mediated transposition: transposition to a target site → this is followed by a loss or retention of the transposable elelment at the donor site
( C ) retrotransposition: the element is transcribed into RNA which is then reverse-transcribed in to cDNA → the cDNA copy is either inserted or copied into the target site
TEs are not always neutral → they can lead to gene reverse transcription and random insertion
Neutral theory of genome evolution
theory: genetic drift may be important in shaping genome features → Repetitive DNA, introns, TEs, and large tracts of non-coding DNA → may have accumulated not because of adaptive processes but rather the inability of selection to work
smaller organisms = large Ne
larger organisms = smaller Ne
lower recombination rate and higher drift may lead to larger genome sizes → Multicellular organisms more likely to
accumulate repetitive DNA (introns, transposable elements, and large tracts of non-coding DNA) through genetic drift →Transition to multi-cellularity led to
- lower recombination rates
- stronger drift
→ This increases the likelihood of mildly deleterious mutation accumulation → This reduces the efficacy of selection
→ Endosymbiotic bacteria accumulate mildly deleterious mutations
Recombination rate
larger number of chromosomes = higher recombination rate
lower number of chromosomes = lower recombination rate
multicellular organisms are more likley to accumularte repetative DNA
transposable chromosomes have a higher recombination rate
Orgins of new genes
histones: proteins with DNA wrapped around them
the regulatory regions is upstream of the coding regions
alternative splicing: different versions of our mRNA
-→ evolution of new genes is due to nuplication and co-option opf whole developmental pathways
Gene duplication
This is how novel genes arise
Subfunctionalization: functions become subdivided among copies
Neofunctionalization: novel functions arrive
orthologous genes: homologous genes in two or more species —> split copies of the duplicate genes
paralogous genes: homologous genes (gene copies) that are related by gene duplication → 2 new duplicated versions of genes
→ two major mechanisms of gene duplication:
unequal crossing over: chromosomes are incorrectly alligned → this leads to tandem duplicates
retrotransposition of mRNA → loss of introns in the copu
→ most duplicated genes are deleterious and so are non-functional (dead on arrival)
changes in gene dose
retrotransposition
→ color blindness - result of gene duplication
→ majority of gene duplications are lost → only a small fraction that reach fixation are quickly pseudogenized
→ most gene duplicationsa re selected out of the population immediately lost and only a SMALL amount stick around
Paralogs that are not lost and sbsequentially diverge may have 2 fates:
suibfunctionalization
neofunctionalization
Subfunctionalization
both paralogs may diverge in sequence and in function
both paralogs may lose complementary components of the ancestral function → this may reduce the pleiotrophy of both paralogs
Neofunctionalization
the second copy has a totally new function
this is an example of convergent evolution
Cop
Copy number variation
a type of genetic variation where sections of the genome are repeated or deleted, leading to differences in the number of copies of a particular DNA segment between individuals.
Gene Chimerism
happens when a single gene (or transcript) is formed from two originally separate genes or gene fragments → encodes a hybrid protein with domains from different sources → It can be beneficial in evolution (new functions) or harmful in disease (like cancer fusions)
exon shuffling: new chimeric genes originate through novel combinations of exons from non-homologous recombination between genes
domain: small segment of proteins with specific functions
hypothesis: many genes evolved though exon shuffling between genes
Horizontal gene transfer: genes transferred from distant lineages
Inversions
happen when a large chunk of the chromosom gets swapped or flipped → same gene but the orger is inverted in the chromosome → this creates new functions and regulatory programs → help lead to different phenotypes, mating, and bahavior (chromosome loops happen too)
example: bird ruff
Selective sweep
definition: positive selection acting to fix a newly arisen advantageous mutation
genetic diversity is reduced at the site of an advantageous mutation that have undergon selective sweep
three factors that interact to determin ethe amount by which diversity is reduced at or near a selection locus:
strength of selection
recombination rate
the time since the selective sweep
The selection sweep:
stronger selection makes the stretch of diversity wider
stronger selection leads to a reduction in diversity over larger genomic regions (makes the stretch of reduced diversity wider)
How much the beneficial allele increases fitness compared to alternatives
Strong selection (large s): allele rises to fixation fast → sharp reduction in genetic diversity
Weak selection (small s): allele rises slowly → more time for recombination and drift
Recombination rate:
Lower recombination leads to a reduction in diversity over larger genomic regions
haplotype: a set of alleles that physically link on a chromosome
new beneficial allele arises on haplotypes → this happenes to be a rare recombination → after the sweep, genetic variance is still reduced but not as strongly
heyterozygosity is reduces in regions of low recombination
How often crossing-over breaks linkage between the selected allele and nearby neutral alleles
Low recombination: large “hitchhiking” region → big chunk of genome around the beneficial allele shows reduced variation
High recombination: sweep effect is localized → only the immediate neighborhood of the selected site shows reduced diversity
Time since the selective sweep
Definition: How long ago the beneficial allele fixed
Effect on sweep:
Recent sweep: strong signature remains → reduced diversity, extended haplotype homozygosity (long stretches of identical DNA).
Old sweep: recombination and new mutations restore variation → signal becomes faint or erased over timeMutation arises -< advantageous mutation increases in frequency → later it is fixed and all chromnosomes coalese soon after it appears
This is the patterns of how many individuals/populations carry the mutation and the patterns of genetic linkage to indicate the history of that sweep
—> summary:
Strong selection = faster, more dramatic sweep.
Low recombination = larger genomic region affected.
Recent sweep = stronger detectable signal.
Hitchhiking effect
This is the indirect impact of selection on patterns of linked diversity
a new beneficial mutation arises on a particular genetic background (haplotype) among chromosomes in a population and after a few generations → the most fit genotyupe has the most offspring → happens if there is no recombination
reduces genetic diversity
One beneficial afllele can sweep to fixation
When a beneficial allele increases in frequency due to positive selection, the nearby neutral (or even slightly deleterious) alleles that are physically linked to it on the chromosome also rise in frequency → carried alomg the ride
Creates long blocks of haplotype homozygosity
Creates long blocks of haplotype homozygosity
Soft Sweeps
maintains genetic variations
beneficial allele is present in multiple copies in the population before selection
a few mutations that are close to each other and are both beneficial, and it takes a lot of time for these beneficial mutations to recombine before they add a lot of fitness to the genotype = more diversity in the genotypes that get swept

Hard sweeps
A single new mutation arises that is beneficial and sweeps to fixation
neutral alleles dragged to fixation → the new advantageous mutation sweeps through a population very quickly → by the end of the sweep, everyone single individual has the exact same mutation

Background selection (reverse hitchhiking)
decreases neutral genetic polymorphism
the reduction of genetic variation at neutral sites due to purifying (negative) selection removing deleterious alleles from the population
decreases genetic diversity
stronger selection leasds to reduction in diversity over larger genomic regions
Gene Flow
this is the mix of alleles from different populations (aka migration)
new alleles are introduced into the population
homogenious allele frequencies across populations
results from dispersal = the movement of individuals and/or gametes
→ measuring gene flow:
if populations are discrete → m = migration rate = the fraction of individuals in a population that are migrant
→ how gene flow homogenizes allele frequencies:
Change in frequency = migration rate (m) (frequency in source population → where the migrants come from (pm) MINUS the frequency in the the present population → where the migrants have gone to (p))
new p = present population MINUS the change in p
B
Barriers to gene flow permit diffrientiation (and drift)
lower rates of miigration and/or smaller populations sizes increase drift rates
Fst → measures genetic diffrientiation among populations → if Fst is higher → then there is MORE genetic differentiation among populations (ranges from 0-1)
Fst = 1 → a lot of genetic variation
Fst = 0 → no genetic variation between the 2 population (more similar)
used to identify genes underlying adaptive traits
this leads to Isolation by distance: Fst increases smoothly with physical distance between populations → further away means less likely to be more genetically related higher Fst value (Fst value is porportional to the distance → higher Fst = more distance)
-→ Isolation by distance (IBD) = MAIN driver of differences between human populations
Linkage disequilibriums (LD)
when an allele at one locus is found together with an allele at a second locus in a population more often than expected by chance → no independent assortment → occurs when alleles at two (or more) loci are associated more (or less) often than expected by chance → knowing the allele at one locus gives you information about the allele at the other locus
LD is influenced/caused by:
Recombination: breaks down LD over time → the higher it is the faster LD erodes → takes more than 1 generatrion
Selection: can increase LD if certain allele combinations are favored.
Genetic drift: random sampling can create or maintain LD in small populations.
genetic hitchhiking: may lead to LD
non-random mating: can maintain LD
Heterostyly prevent inbreeding
Linkage equilibrium
alelles are inherited independently
Sexual recombination
asexual → no recombination
sexual → with recombination
What is sex?
sexual reproduction is the fusion of 2 gametes and the subsequent segregation of chromosomes by meiosis
usually accompanied by recombination
it is very diverse:
hermaphroditic (can change to male or female sex )
sequential hermaphroditism (male to female)
cleisstogamous
conjugation
High costs of Sex
finding a mate: energetically costly and dangerous (STIS and vulnerability)
can break up winning genotypes →recombination can make new combinations of alleles that are worse or bad
two fold cost of sex →
sexual organism produce both males and females → only females produce offspring
ASEXUAL organisms only produce females → every offspring can reproduce → many reproducers
Cost of males: sexual organisms pass on ½ of their genome
In sexual populations, males don’t produce offspring themselves, so half the population (males) is not directly contributing copies of their genome to the next generation → every individual reproduces, passing on their entire genome
asexual organisms pass on whole genomes → genome fully inherited
in sexual populations, genes are diluted
Asexual reproduction
colonal reproduction may only last for a few generations
asexual species are associated with invations → Reproductive advantage (no mate needed) → A single individual can establish a new population because it doesn’t need a mate to reproduce → causes rapid population growth
the entire genome has the same gene tree and is in LD since there is no recombination and they got their genome from colonial inheritance → high LD
Advantages of Sex
new combinations: suffling of alalles during recombination creates novel genotypes → some may be asaptive
efficient selection: recombination breaks up LD between beneficial and deleterious alleles
combine beneficial mutations: recombination allows beneficial; mutations to be combined ina single individual→ in asexual reproductionm the alleles are in compettion
Removed deleterious mutations: recombination allows deleterious mutations to be removed
→ advantage to sex = RECOMBINATION!!
Disadvantage to asexuality
asexual species accumulate deleterious mutations
hitchhiking of deleterious mutations
due to mullers ratchet
asexual species adapt slower
forced to fix advantageous mutaions sequentially
creation of novel haplotypes in asexuals is limited by the rate of mutations _> makes it hard to keep pace with rapidly evolving pathogens (red queen hypothesis)
Muller’s Ratchet
talks about mutation accumulation → ONLY works in asexual organisms
in asexuals → process in asexual populations where deleterious mutations accumulate irreversibly over generations
no recombination → once the best or “least mutated” genotype is lost by genetic drift → you cannot recreate it or get it back
this lowers the mean fitness
in sexual organism → mullers ratchet is avoided since recombination can reform the haplotypwe that lacks deleterious mutations
Experiemental evidence for the accumulation of deleterious mutations in asexual populations → yeast mutation trajectories
Sexual yeast removed deleterious mutations and only the beneficial alleles made it in the population
Asexual organisms accumulate more deleterious alleles
Higher number of dN/dS
Colonial interference hypothesis
without sex/recombination → selected alleles must fix sequentially
WITH sex/recombination → selected alleles can fix simultaneously
the max rate of adaptation is limited in asexual specieies
In asexual species: combinations of alleles are not formed until a second mutation occurs in a lineage that already has a first mutation
in sexual populations: independent mutations are brought together un a lineage more rapidly by recombination → adaptation is more rapidly achieved
The red queen hypothesis
to stay the same fitness you need to be constantly adapting to the changes in your environment → recombination allow sexual species to evolve more rapidly to resist parasites
Sex is an advantage in constant co-evolutionalry arms races between hosts and parasites
Arms race with parasites/pathogens
What are sexes
anisogamy: gametes of different sizes
male: small and mobile gamtes
females: larger and less mobile gametes that provide maternal provisioning (nutritional input)
→ why have seperate sexes arose?
selection for specialization
inbreeding avoidance
secondary sexuyal traits → showy or exagerated traits
Sexual selection
diffferences among individuals in their reproductive success causes competition for access to mates or fertilization opportunitues
fitness = fucunbdity * viability
diffrient traits or selective pressures trhat uncrease fucundity
Sexual dimophism
viability selection may favor the reduction of showy traits in both sexes → but sexual selection can favor a stronger increase on one sex
exagerated traits = better reproductive success → can find mates better
How sex is determined
some genes have hermaphrodites
haploid, diploid
most plants are hermaphroditic and dont really have a sex determination system
sex determination = involves genes and the environement and also the [hysiology (hormones)
one chromosome may be reduced in size and may not undergo recombination → gets very small
Heteromorphic sex chromosomes have evolved independently many times
How the evolution of sex chromosomes work
they are apart of ancestral autosomes
they evolve a sex determining locus
spression of recombination between neo-X and neo-Y
degeneration of neo-Y due to muller’s ratchet and hitchkling of deleterious alleles
Sexually antigonistic allele
this is an allele that has a benefit for one sex and cost for the other
sex determining allele arises
recombination between sexually antagonistic allele and male-determining allele lowers fitness
selection for recombinmation is supressed between these loci
this leads to shutting off recombination and no means that selection of proto-Y no longer recomines
this leads to the degeneration of the Y sex chromosome and the accumulation of repeats and transposable elecments
old chromosomes can be longer due to TEs
Evolutionary strata
specific chunks of our X and Y chromosomes stopped recomning due to differnt inversions and transpositions at specific times in history of our genus and species
most decay chromosome size = oldest strata → inversion of the Y chromosome
halting recombination leads to the decay of y chromosomes
Inversions can lead to the sudden reduction of recombination
Bateman gradient
used to measure the strength of selection
fertility gradient of multiple matings can differ
this tells us the variation between individuals in males and females
thisalso tells us the selection gradient between fertility and number or mates or mating events
and tells us the variance in reproductive success and how it can differ between species
Bateman gradient measures how strongly an individual’s reproductive success increases with the number of mates
males usually have a steeper gradient slope since → stronger sexual selection:
Males: Reproductive success increases steeply with number of mates.
Each additional mating can directly add more offspring (since sperm is cheap and plentiful).
Females: Reproductive success increases much more slowly (or not at all after the first mate).
Egg production is limited, so having more mates doesn’t usually give more offspring.
Evolution for sexual dimorphism
females are more showy
males are more choosy
males suffer less by mating with an inappropriate female but females’ fitness is significantly lowered if they mated with the wrong male since their developing sygot will almost always impacts the female parent fitness
maternal parent prodive more or most of the care for their offspring
Measuring difference in slection pressures
I = the opportunity for selection → differnce of variance in fitness (number of offspring) between in males and females (males MINUS females = difference or change in these 2 measurments) → used to decide who has more variance in the offspring they have
males often have great variance in reproductive success
when males take care of their offspring then the females are more showy ( not very common but is still present)
Why it pays to be showy
intrasexual selection: competions between individuals of the SAME SEX for mating and fetelization opportunities →WITHIN one sex
intersexual selection: selection due to the choice by the OPPOSITE SEX → BETWEEN
Intrasexual selection
intrasexual selection: often male-male competitions
post copulatory intrasexual selection:
adaptations to ensure fertilization
removing rivals sperms, mucus plugs, etc
Female choice: bias towards males with particular chacters:
direct benefit
indirect benefits
good genes
fisharian runaway selection
sensory exploitation
Direct benefits by mate choice
Help raising offspring
Territory
“Nuptial” gifts
Greater fertility
Even if mates provide nothing there can still be direct benefits → spotting males from afar without having to wonder around getting eating by a predator looking for a mate and showy traits help determine with individual is healthy or have no parasites
Indirect benefits of intrasexual selection
good genes hypothesis
male has female trait that attracts females and vis versa → caused by assortive mating
Fisherian runaway sexual selection
attractive dads make attractive sons
posotbe feedback look
Initial variation:
Some males have a slightly longer tail, brighter feathers, louder calls, etc.
Some females prefer those traits.
Female choice spreads:
Females who prefer the trait mate with males that have it.
Their sons inherit the trait, and their daughters inherit the preference.
Now the trait and preference become genetically correlated.
Runaway process:
As both the preference and the trait spread, the trait becomes more and more extreme.
This “runaway” continues until the survival cost balances the mating advantage
Sex ratio
usually 1:1 or 50/50
when the sex ratio balance is off, some animals can change their sex to revert the balace back
optimal sex ratio = more females than males → cost of males = disadvantageous to sexual reproduction
thisis an example of evolutionary stable strategy (ESS)
for an autosomal allele → 50/50 sex ration is an ESS but for an allele on a sex chromosome it is not → Y can only be passed to YX offspring
Sex ratio disorders
X chromosome of a father is not transmitted to his male offspring → therefore the X chromosome sin the males can benefit by evolving to damage or kill sperm carrying the Y → winters sex ratio distorter → after it preads → suppression of distorters can arise and spread → thi restores the sex ration back to 50/50
Autosomal killer systems (damage sperm or eggs) → less common → Chromosomes make extra centromeres on chromosome, → makes it more likely that a microtubule attaches and pulls the allele down → female gametes only the last bottom gamete survive → making these extra centromeres allows the gamete to have a higher survival rate -→ can kill all the eggs or all the sperm
Kin selection
self sacrificing behaviro that benefits relatives
inclusive fitness: it measures the transmission of alleles that are IBD and increases if individuals are alturistic → this is the idea that the fitnes is made up of direct+ indirect fitness
B = beneit
C = cost
r = degree of relatedness between actor and recipient
need to follow Hamilton’s rule to know that they havea shared relationship
eusociality: offspring are nearly or completely sterile workers
Conflict and cooperation
conflict between siblings → bird can bully their sister out of the nest to gewt the full benefits from their parents → occurs when cooperation is not beneficial
Mother-offsping conflict: resource conflicts during pregnancy → fetus may try to keep mother’s blood glucose levels high to get more nutrients → too much nutrients = invasisve placental growth → can lead to fetal overgrowth and/or hut/kill the mother