Training Strategies and Augmentation Techniques for GNNs
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
Reasons and approaches of graph manipulation
Feature level:
The input graph lacks features → feature augmentation
Certain structures are hard to learn by a GNN (e.g. cycle count) → we add them as features
Structure level:
The graph is too sparse = inefficient message passing
→ adding virtual nodes/edgesThe graph is too dense = message passing is too costly
→ random neighbor samplingThe graph is too large = cannot fit the computational graph into a GPU
→ subgraph sampling for embedding computing
It is unlikely that the input graph happens to be the optimal computation graph for embeddings
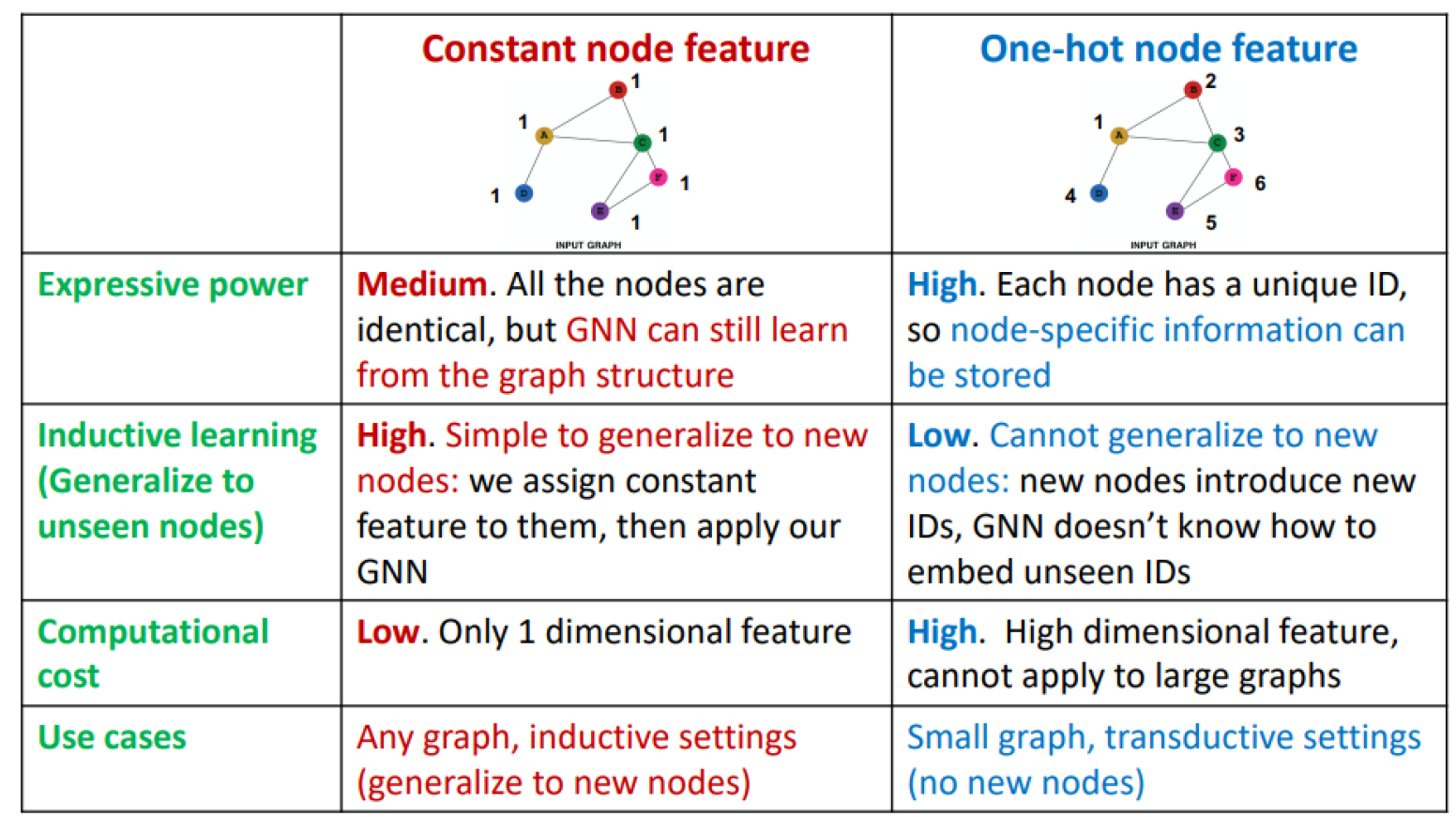
Feature augmentation approaches
Assigning constant values to nodes
Assigning unique IDs to nodes as one-hot vectors
Adding certain features, e.g. centrality, clustering coefficient
Comparison of feature augmentation approaches
Common approach for adding virtual edges
Connecting 2-hop neighbors via virtual edges
→ using A+A2 as computation graph
Use case: bipartite graphs (e.g. author-to-papers graph → 2-hop is author-to-author collaboration)
Method and benefit of adding virtual nodes
The virtual node will connect to all the nodes in the graph → all nodes will have a maximum distance of 2
Benefit: greatly improved message passing
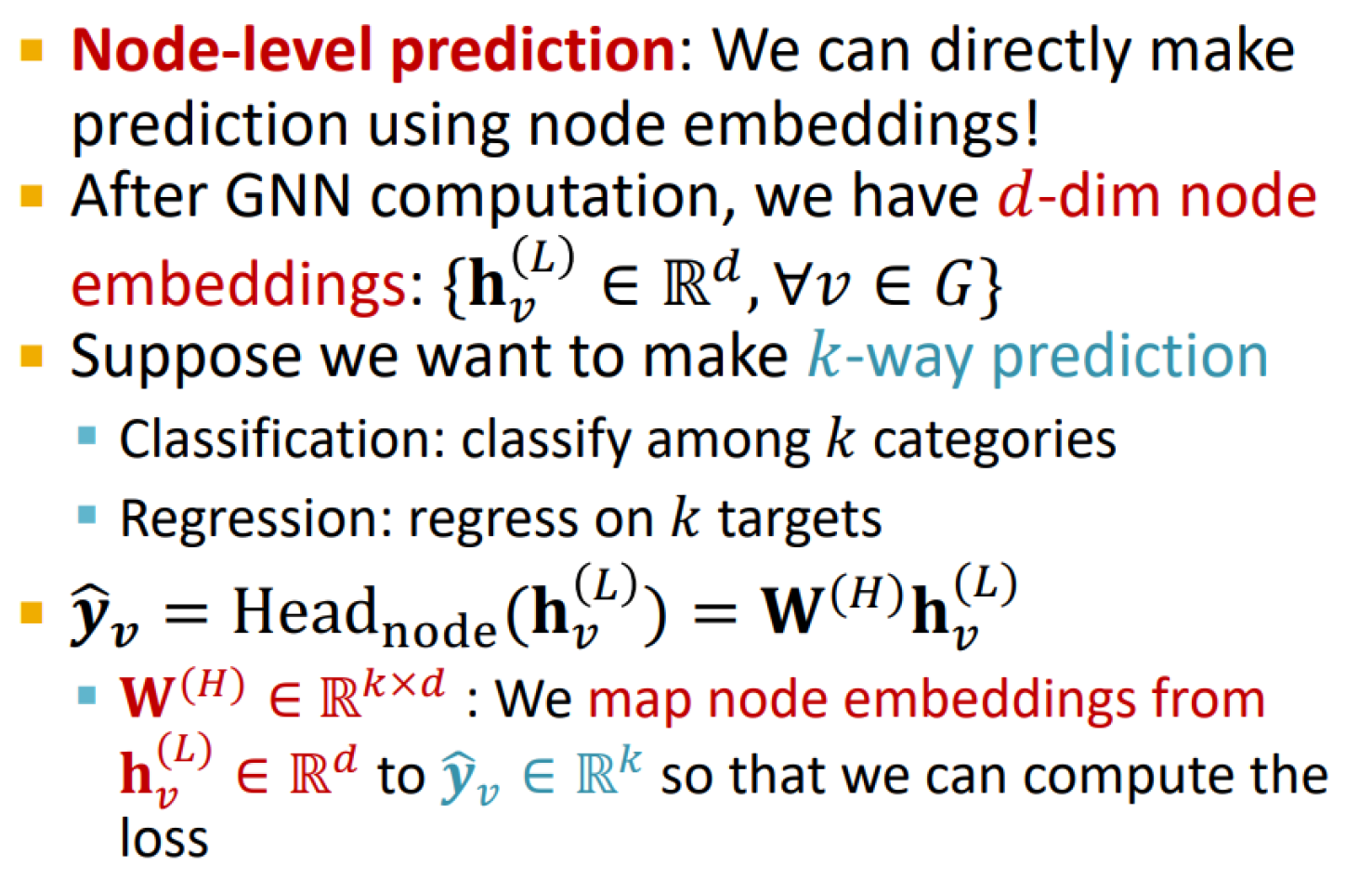
Node-level prediction head
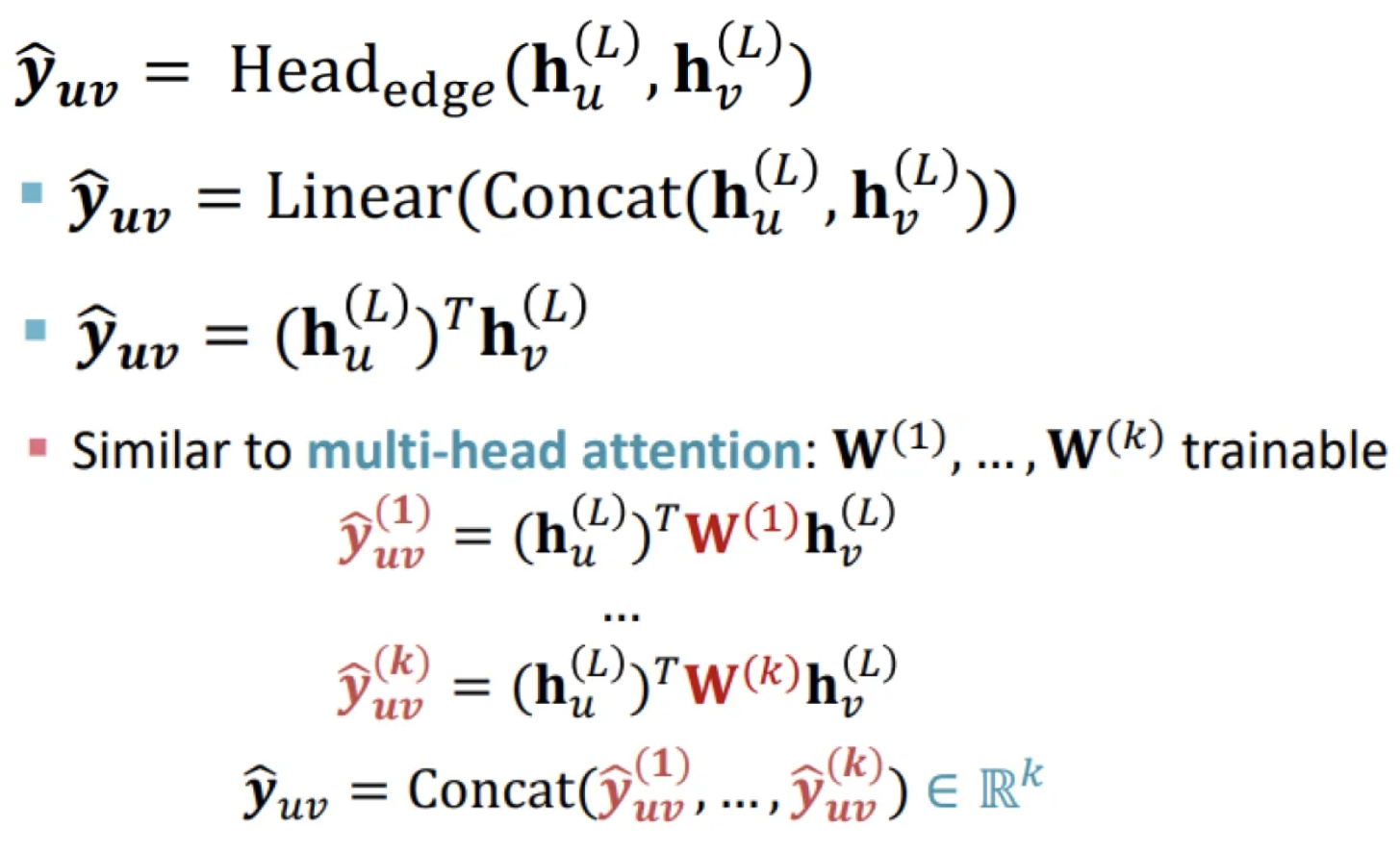
Edge-level prediction head
Make predictions using pairs of node embeddings.
Options:
Concatenation + Linear layer: map 2d-dimensional embeddings to k-dimensional (k-way prediction)
Dot product: simple dot product for one-way prediction, trainable W matrices for k-way prediction
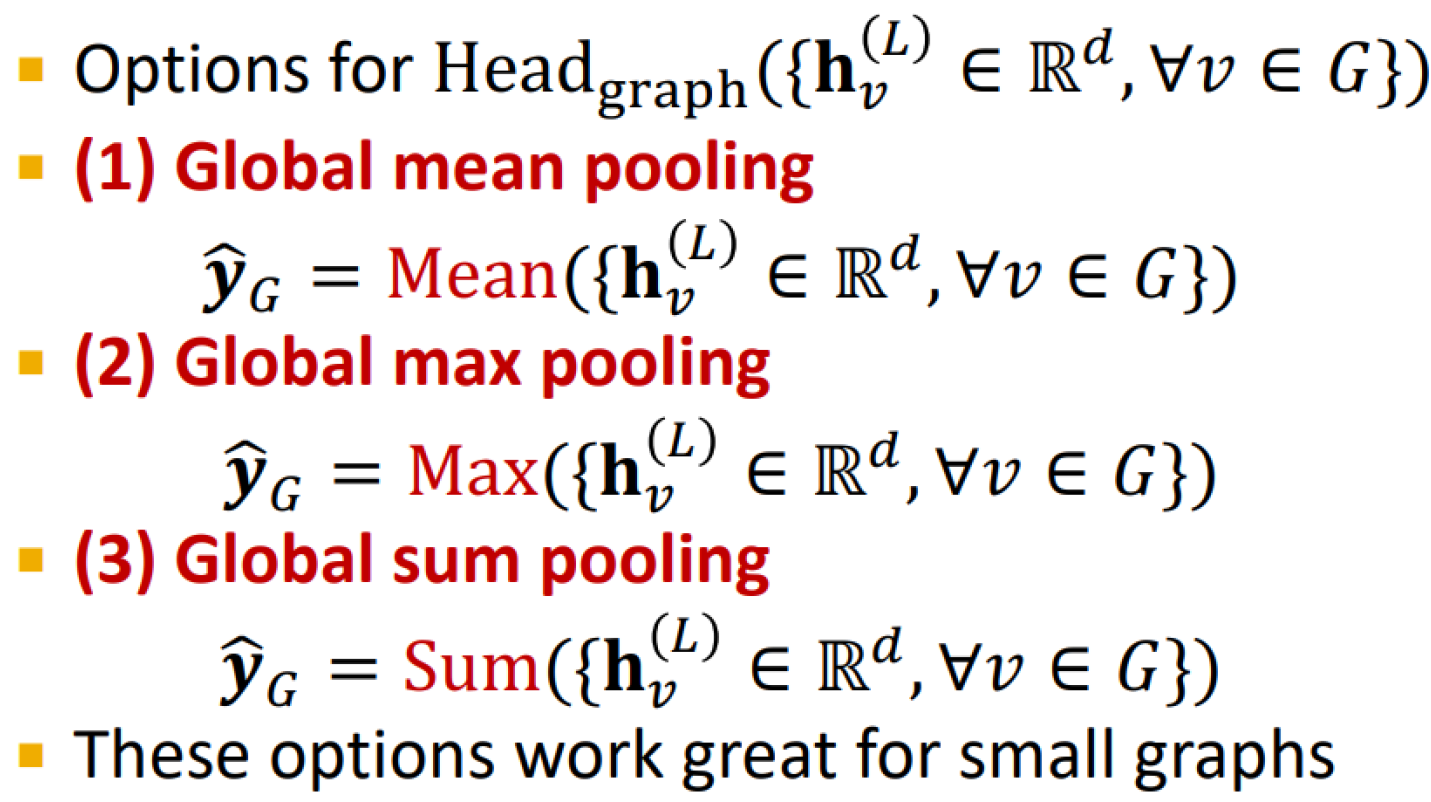
Graph-level prediction head
Make prediction using all the node embeddings in the graph
Unsupervised prediction task examples
Node-level: node statistics, e.g. clustering coefficient
Edge-level: link prediction
Graph-level: deciding isomorphism of two graphs
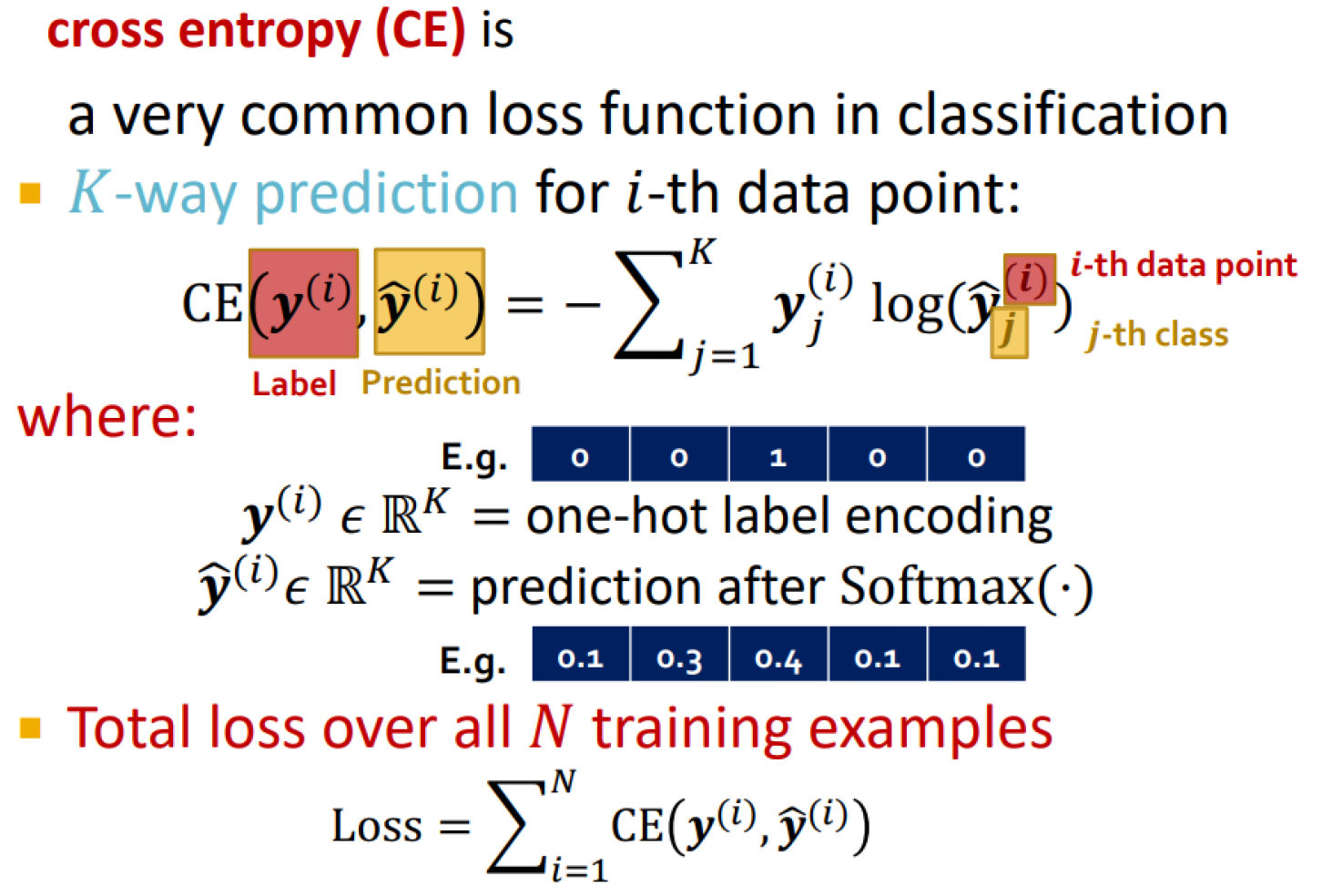
Classification loss
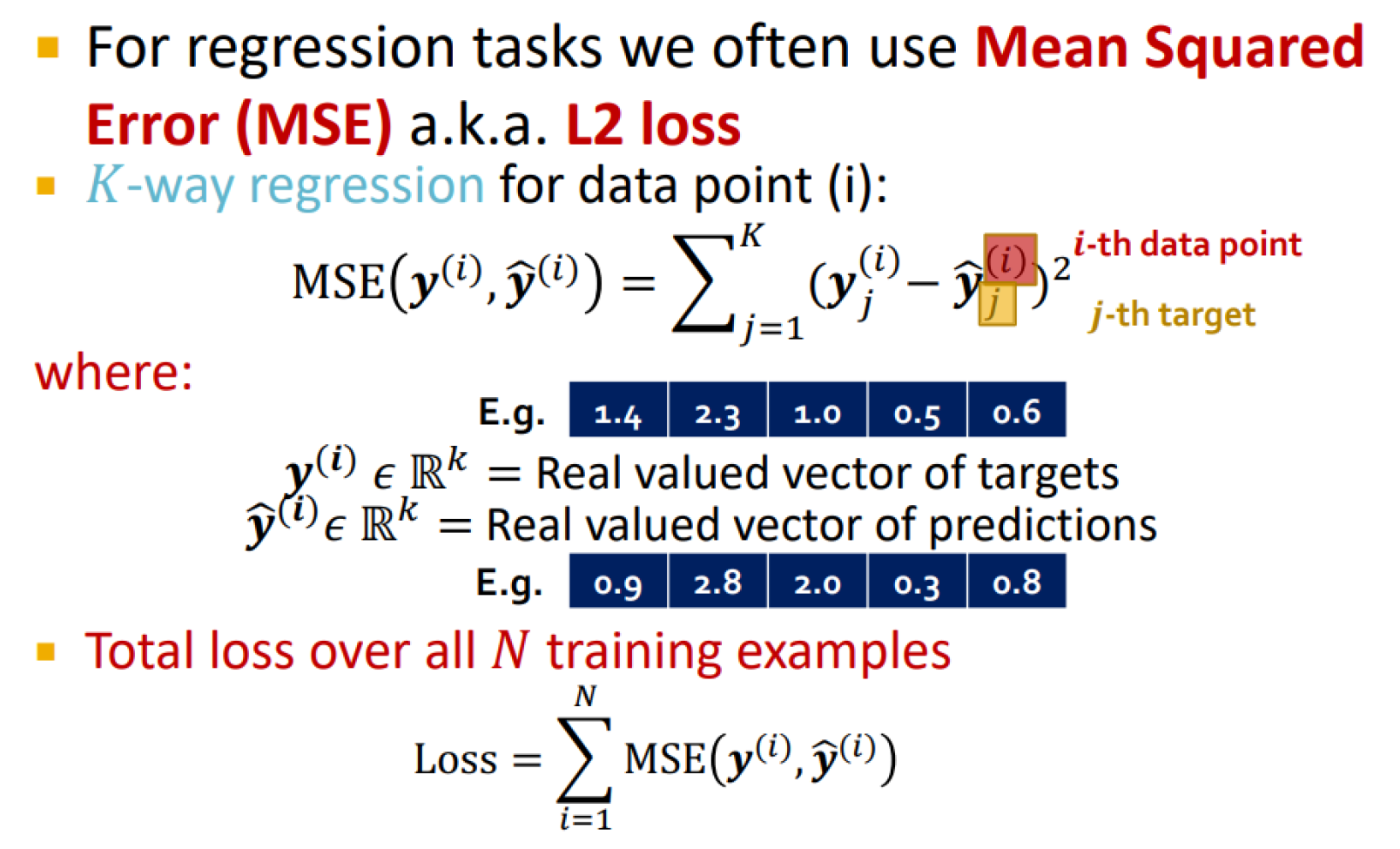
Regression loss
Fixed vs. random dataset split
Fixed: we will split our dataset once
Random: we randomly split, report average performance over different random seeds
Challenge of graph splitting
The nodes (data points) are not independent, they affect each other’s prediction
Transductive vs inductive splitting
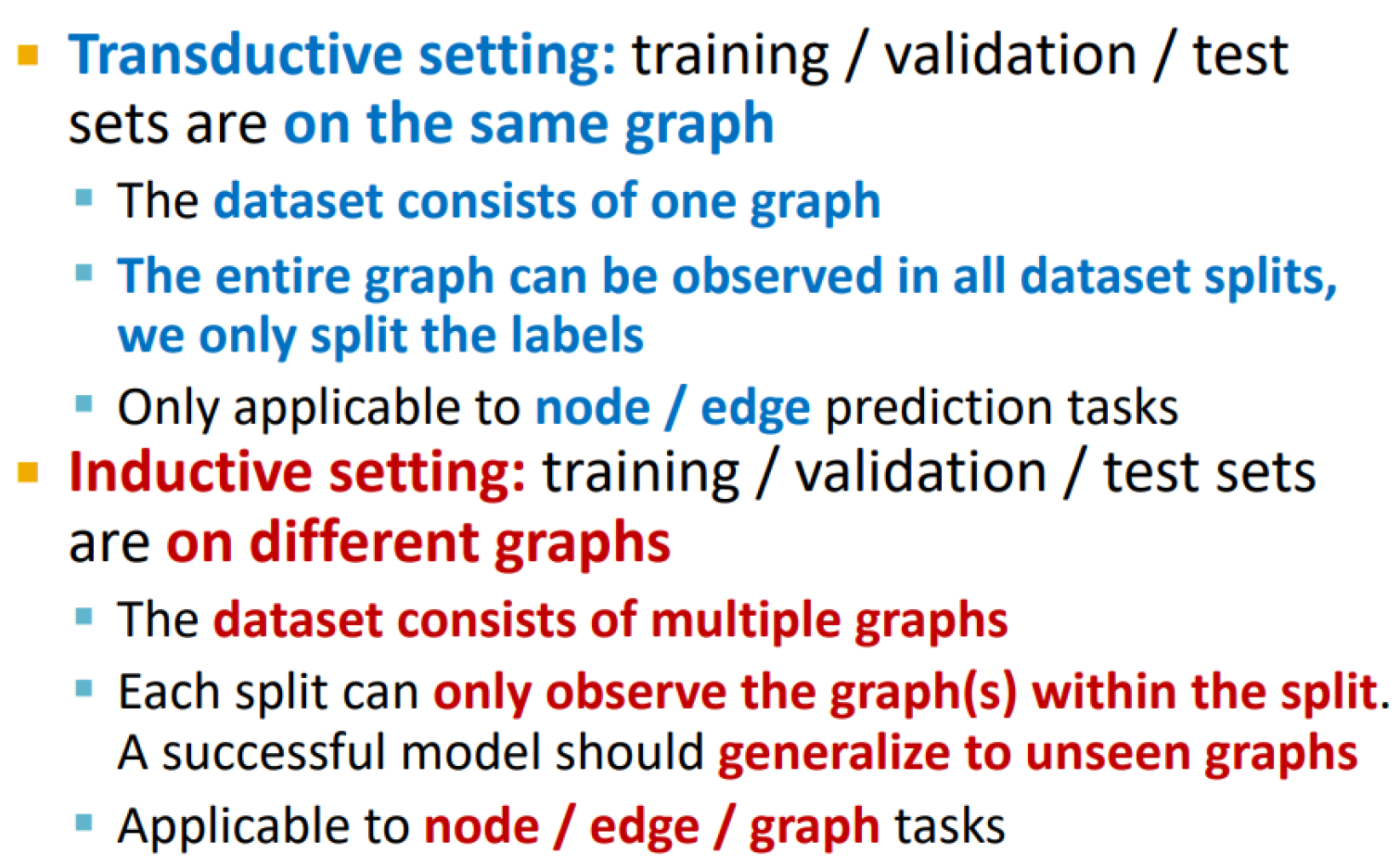
Transductive link prediction
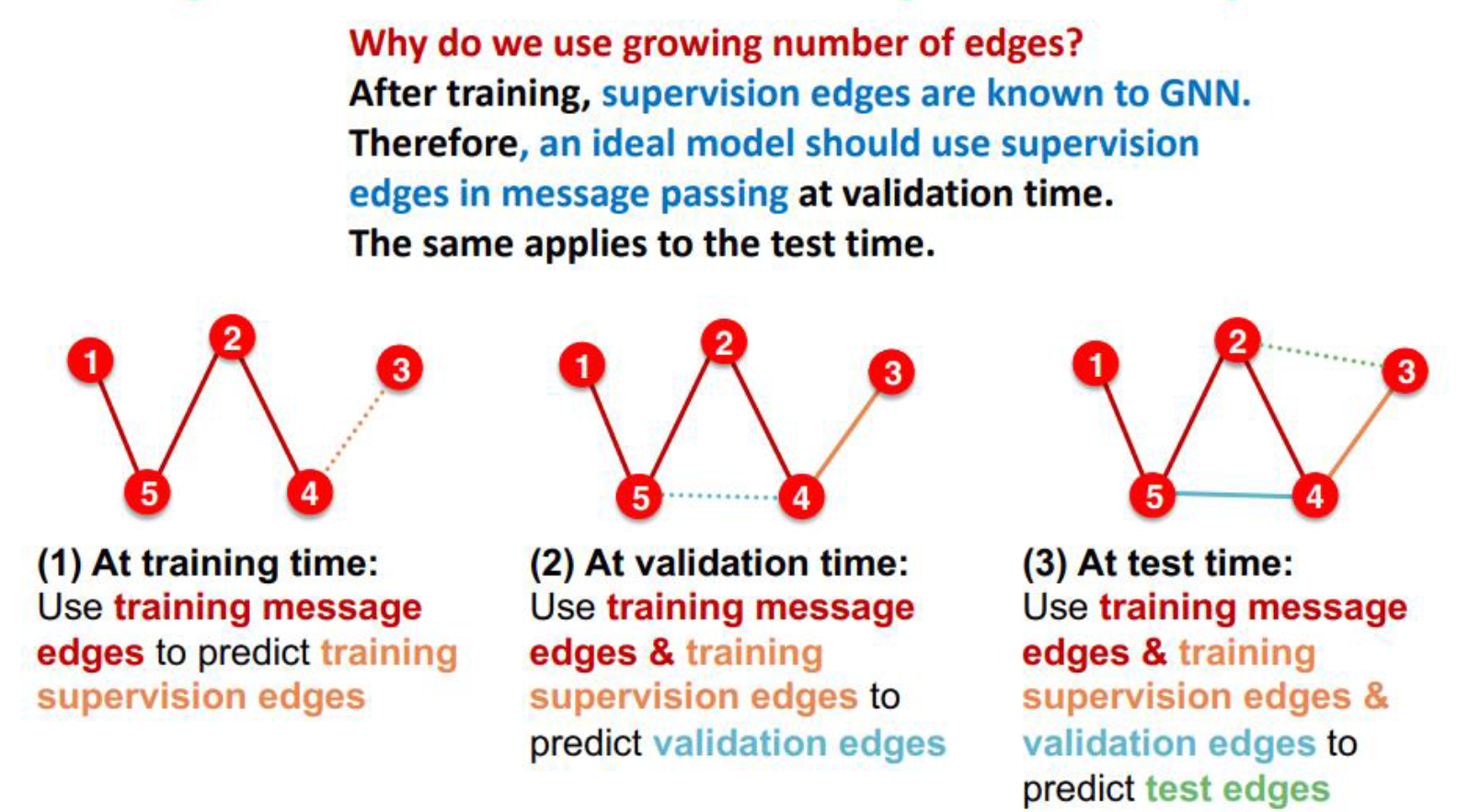
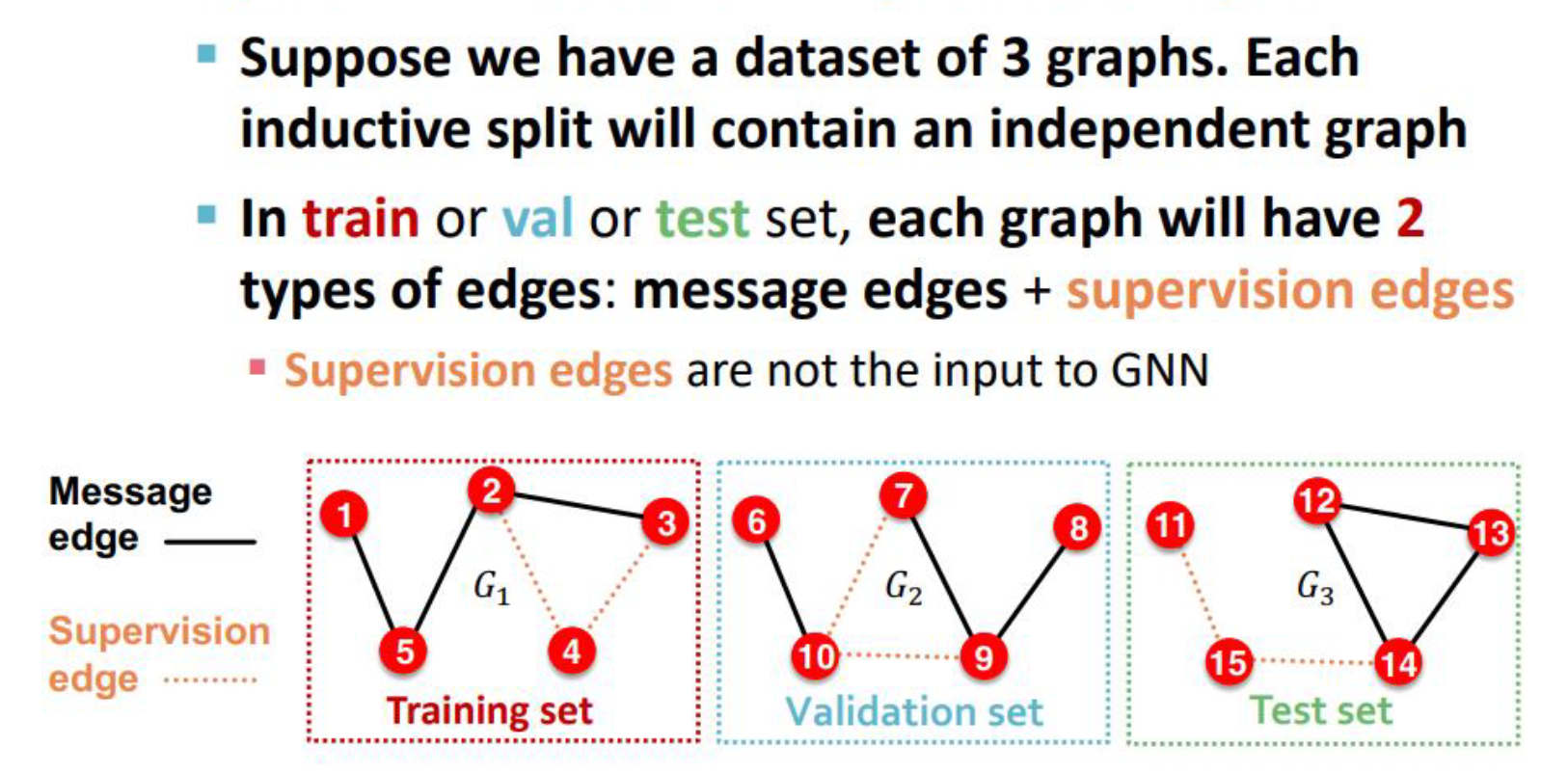
Inductive link splitting
Computational graph and its effect on node embeddings
Computational graphs are defined by the nodes’ neighborhood = rooted subtree structures around each node
→ if two nodes features and computational graphs are the same, the GNN won’t be able to distinguish them, they will get the same embeddings, since it doesn’t care about node IDs
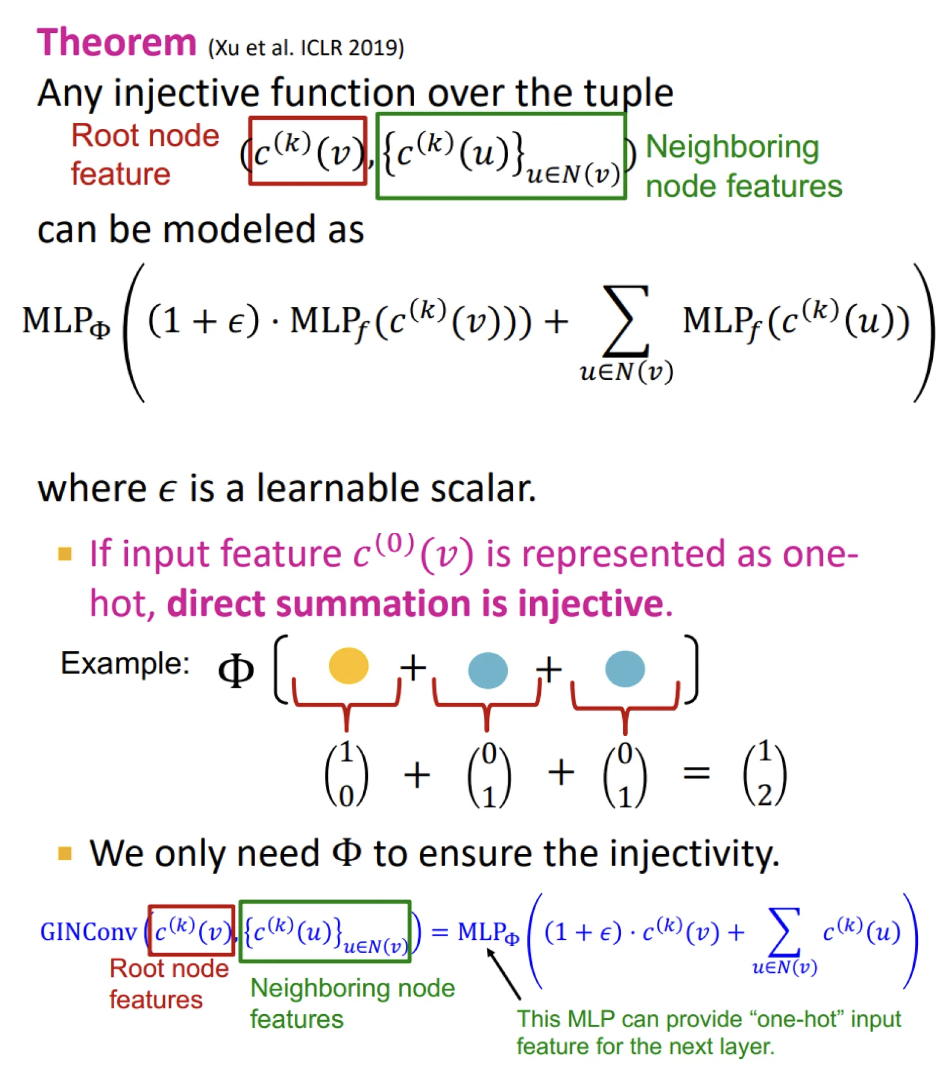
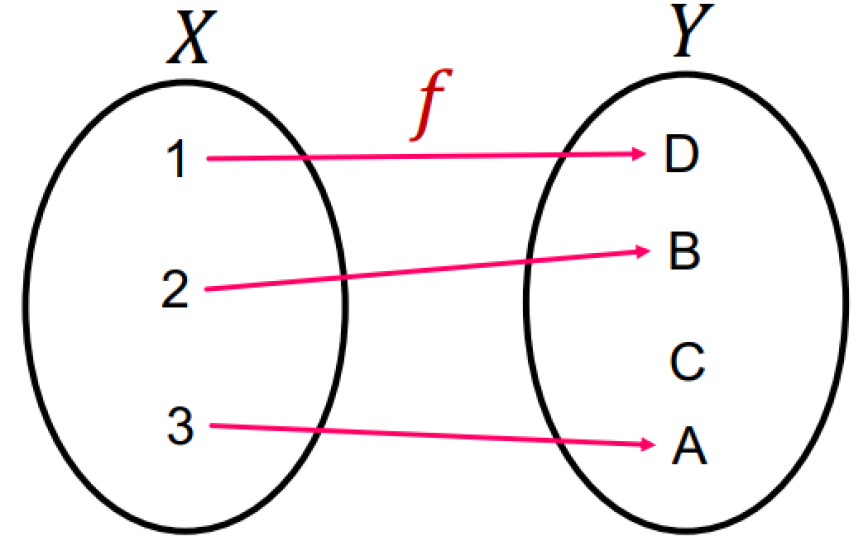
Injective function
It maps different elements into different outputs
→ retains all the information about the input
Most expressive GNN
Maps rooted subtrees to node embeddings injectively.
→ use an injective neighbor aggregation to fully retain the neighboring information and therefore distinguish different subtree structures
Expressive power, GCN and GraphSAGE case
Expressive power of GNNs can be characterized by that of neighbor aggregation function.
Neighbor aggregation is equivalent to a multi-set function
GCN:
element-wise mean pool
failure case: multi-sets with same color proportion
GraphSAGE:
element-wise max pool
failure case: multi-sets with same set of distinct colors
Injective multi-set function
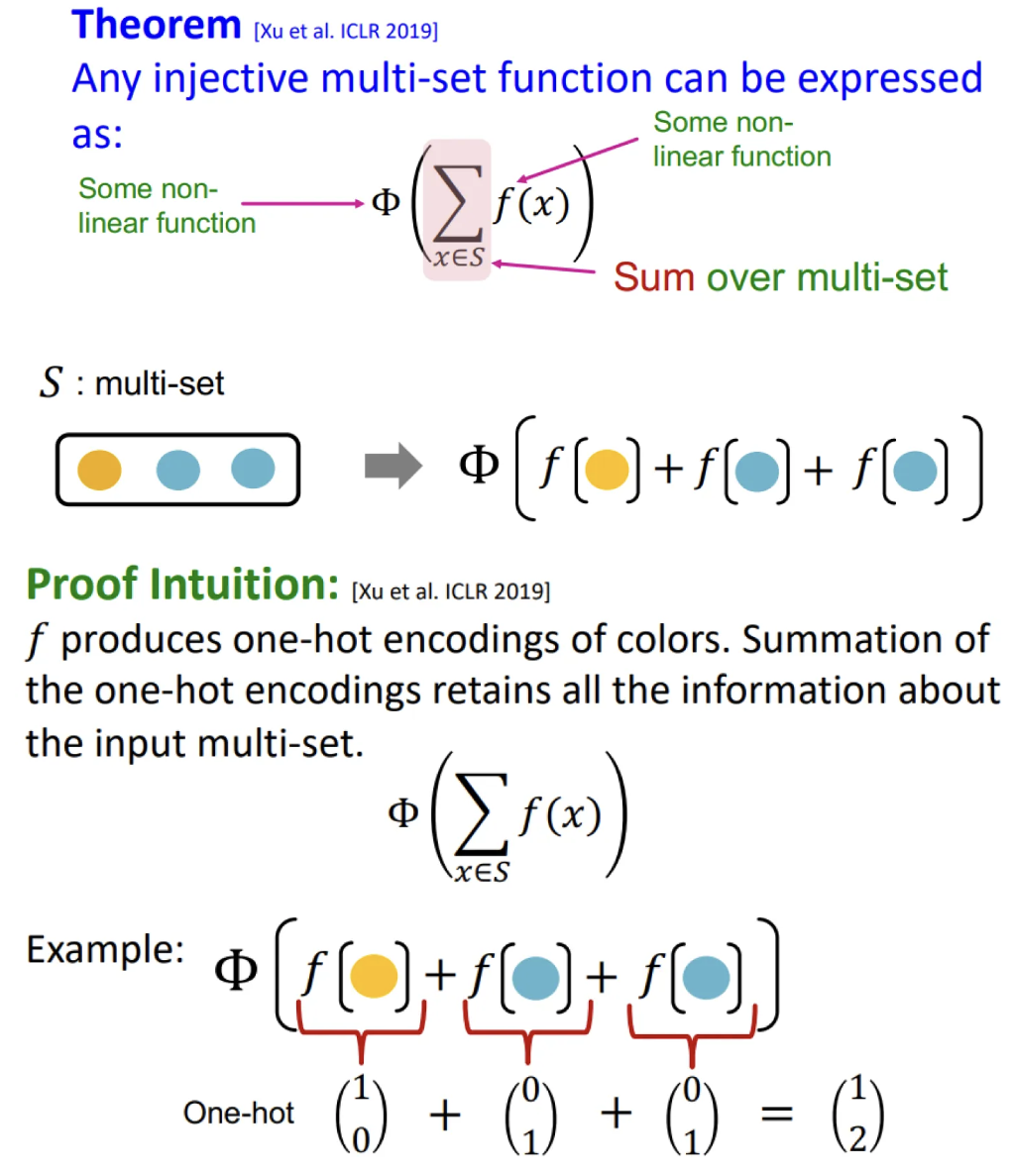
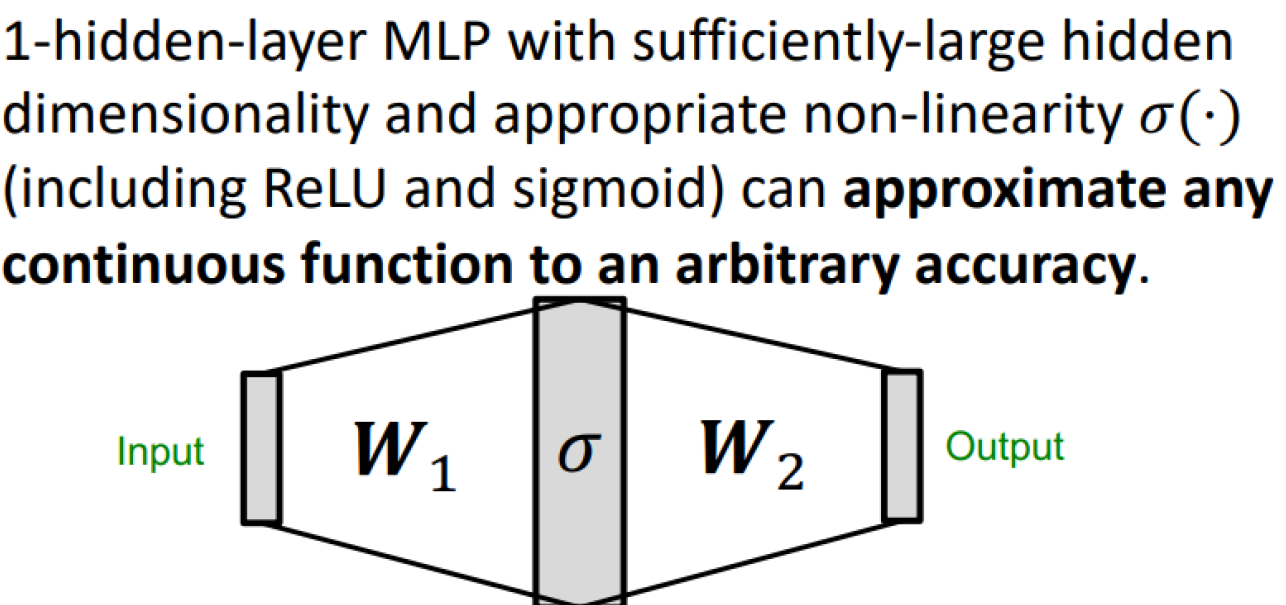
Universal approximation theorem
Graph isomorphism network (GIN)
A differentiable neural network version of the WL graph kernel:
GIN uses a neural network to model the injective hash function
GIN’s update targets are the node embeddings (low-dim vectors), not the one-hot node colors
=> their expressive power is the same, they can distinguish most of the real-world graphs
Advantages:
Due to the low-dimensional node embeddings, they are able to capture fine-grained node similarity
Parameters of the update function can be learnt for the downstream tasks
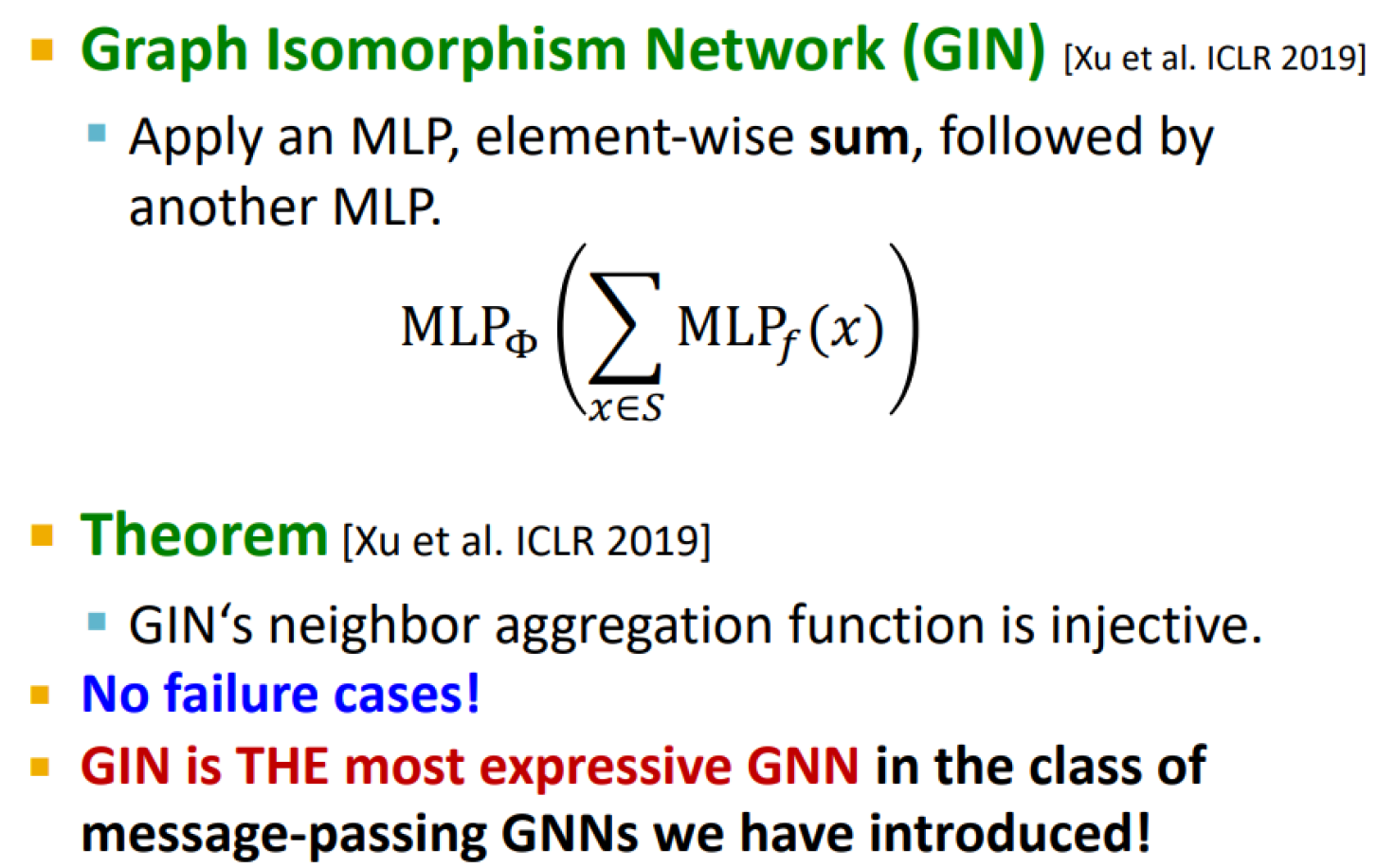
Injective function of GIN
Uses element-wise sum pooling instead of mean/max