Genetics of Microorganisms Exam 2
1/280
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
281 Terms
Gene expression
process by which info from a gene is used to synthesize RNA and polypeptides, and to affect properties of cells and phenotypes
Gene regulation
level of gene expression can vary under different conditions
allows response to environmental changes such as nutrient availability, stress, etc
produce different cells types in multicellular species
facilitates changes during development
Transcription (gene regulation)
Regulatory transcription factors activate or inhibit transcription
Arrangement and composition of nucleosomes
DNA methylation
RNA Modification (gene regulation)
Alternative splicing and RNA editing
Translation (gene regulation)
Proteins regulate translation or mRNA degradation, RNA interference
Posttranslation (gene regulation)
Feedback inhibition and covalent modifications regulate protein function
combinatorial control + 5 factors
most euk genes are regulated by many factors
>1 activator/repressor proteins stimulates/inhibits transcription
activators and repressors modulated by binding of small effector molecules, protein-protein interactions, and covalent modifications
Regulatory proteins alter nucleosomes near promoter
DNA methylation inhibits transcription
Heterochromatin formation inhibits transcription
Transcription factors
proteins that influence ability of RNA pol to transcribe a gene
General and regulatory transcription factors
contain regions called domains
General transcription factors
Required for binding of RNA pol to core promoter and its progression to elongation stage
Necessary for basal transcription
Regulatory transcription factors
Serve to regulate rate of transcription of target genes
influence ability of RNA pol to begin transcription of a particular gene
domains
regions in transcription factors that have specific functions such as DNA-binding, serving as a binding site for small effector molecules, and dimerization
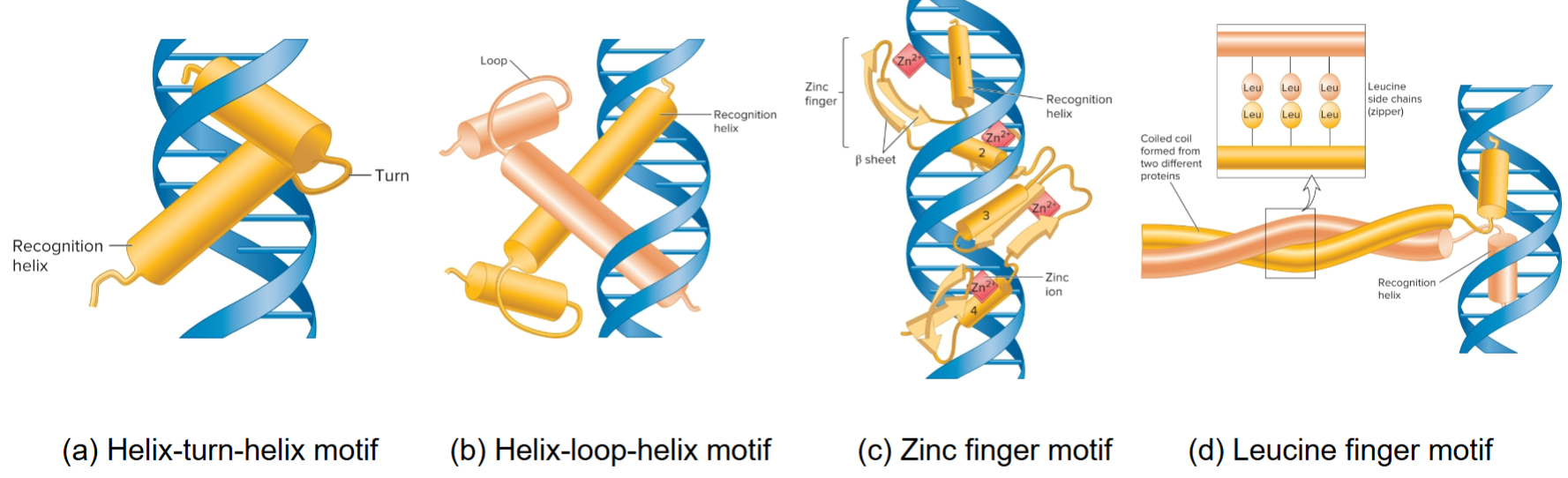
motif
a domain or a portion of a domain in transcription factors that has a very similar structure in many different proteins
control/regulatory elements/sequences
regulatory transcription factors recognize cis regulatory elements within enhancers
binding of regulatory transcription factors to regulatory elements affects transcription of gene
activators and repressors
enhancers
DNA regions (50-1000 bp in length) that contains >1 regulatory elements
binding of an activator to enhancer increases rate of transcription, causing up-regulation which is 10-1,000x
binding of a repressor which inhibits transcription is down-regulation
orientation independent/bidirectional
many regulatory elements within enhancers can function in forward or reverse orientation
regulatory elements can be found within a few hundred nucleotides upstream of promoter
can also be found thousands of nucleotides away, downstream from promoter or even within introns
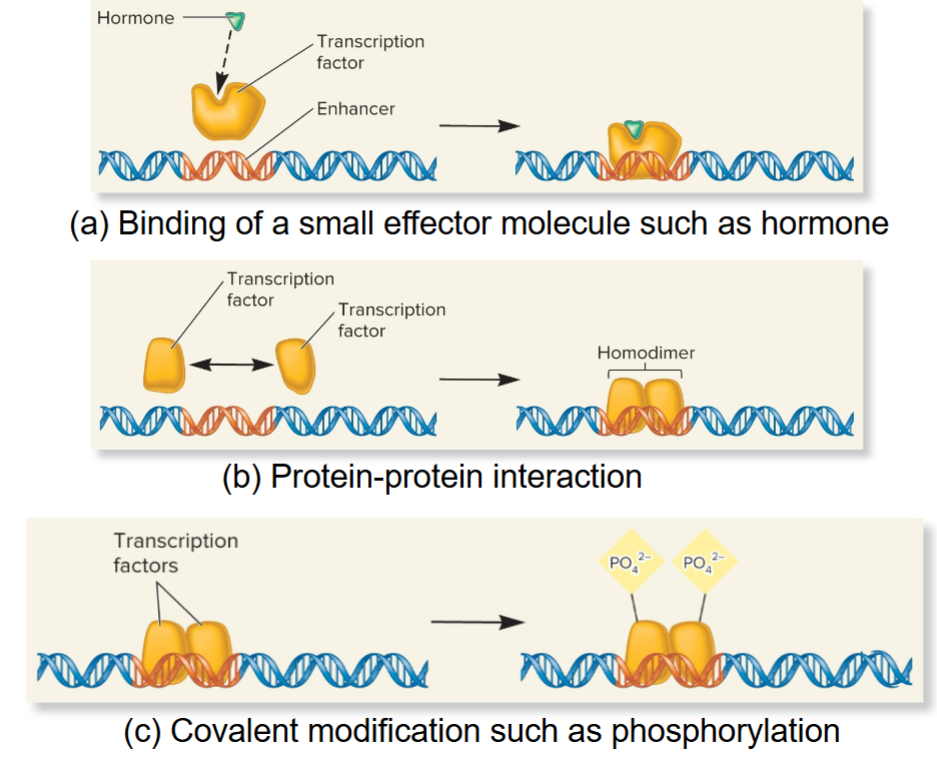
3 common ways function of regulatory transcription factors can be modulated
Binding of small effector molecule
Protein-protein interactions
Covalent modification
open vs closed conformation in chromatin
Chromatin is very dynamic and can alternate between 2 conformations
Open conformation: chromatin is accessible to transcription factors, transcription can take place
Closed conformation: tightly packed, transcription is difficult or impossible
3 things that can affect gene transcription
Positioning of nucleosomes at or near promoters
Presence of histone variants
Covalent modification of histones
ATP-dependent chromatin remodeling
dynamic changes in chromatin structure
nucleosomes change position in cells during gene expression and inactivation
nucleosome positioning changes in promoter region as part of gene activation
transcriptional activators orchestrate changes in chromatin structure
energy of ATP hydrolysis used to drive change in location and/or composition of nucleosomes
makes DNA more/less amenable to transcription
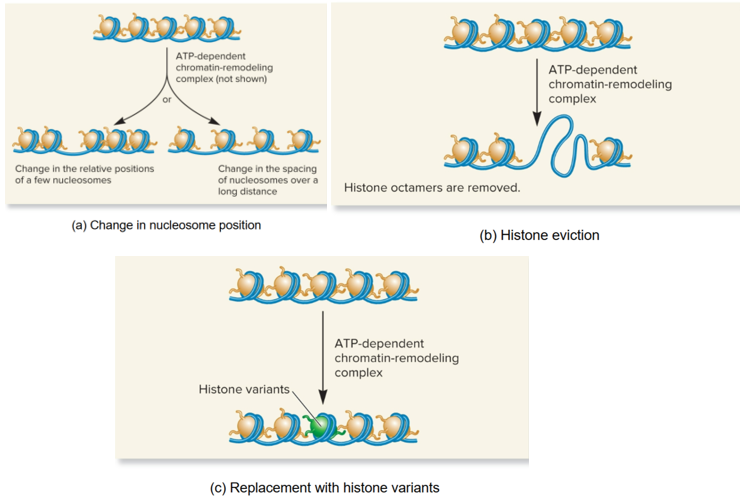
Chromatin remodeling complexes
change chromatin structure by
Change in position of nucleosomes
Eviction of histone octamers
Change in composition of nucleosomes
abnormalities in nucleosomes have been frequently found in cancer cells
DNA translocase
catalytic ATPase subunit that All remodeling complexes have
Eukaryotes have multiple families of chromatin remodelers: SWI/SNF, ISWI, INO80, Mi-2
histones
H1, H2A, H2B, H3 and H4 are moderately repetitive
Human genome contains over 70 histone genes, most code standard histones
A few of these are histone variants which have genes that accumulated mutations that alters AAs
some variants are incorporated into a subset of nucleosomes to create specialized chromatin
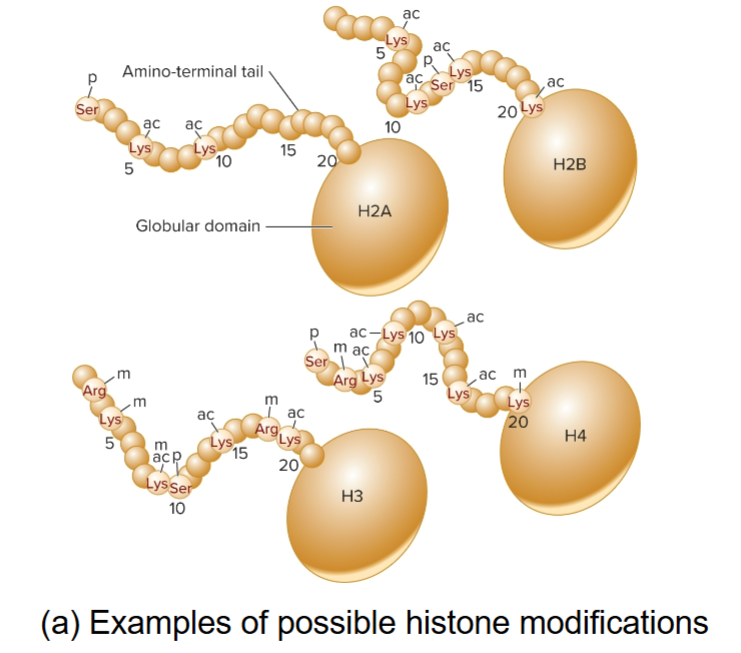
histone code hypothesis
>50 enzymes in mammals that selectively modify amino terminal tails of histones
Acetylation, methylation and phosphorylation affect level of transcription and influence interactions between nucleosomes
occur in patterns that are recognized by proteins
pattern of modifications provide binding sites for proteins that specify alterations to be made to chromatin structure
proteins bind based on the code and affect transcription
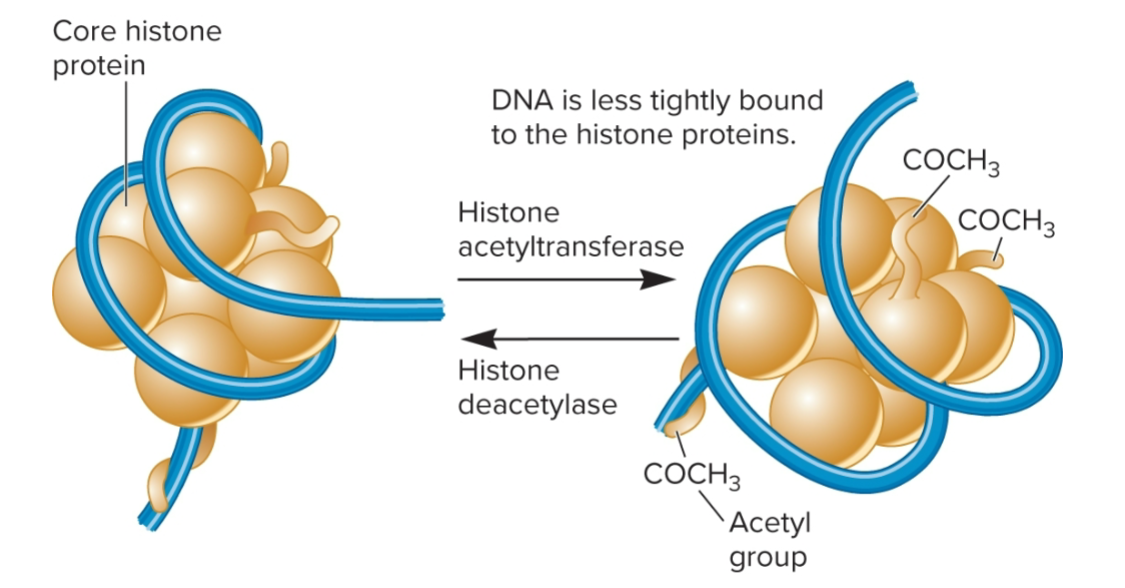
histone acetyltransferases (HATs)
+ charged lysines within core histone proteins can be acetylated
attachment of acetyl group (—COCH3) disrupts electrostatic attraction between histone protein and - charged DNA backbone
Favors open conformation; is highly reversible
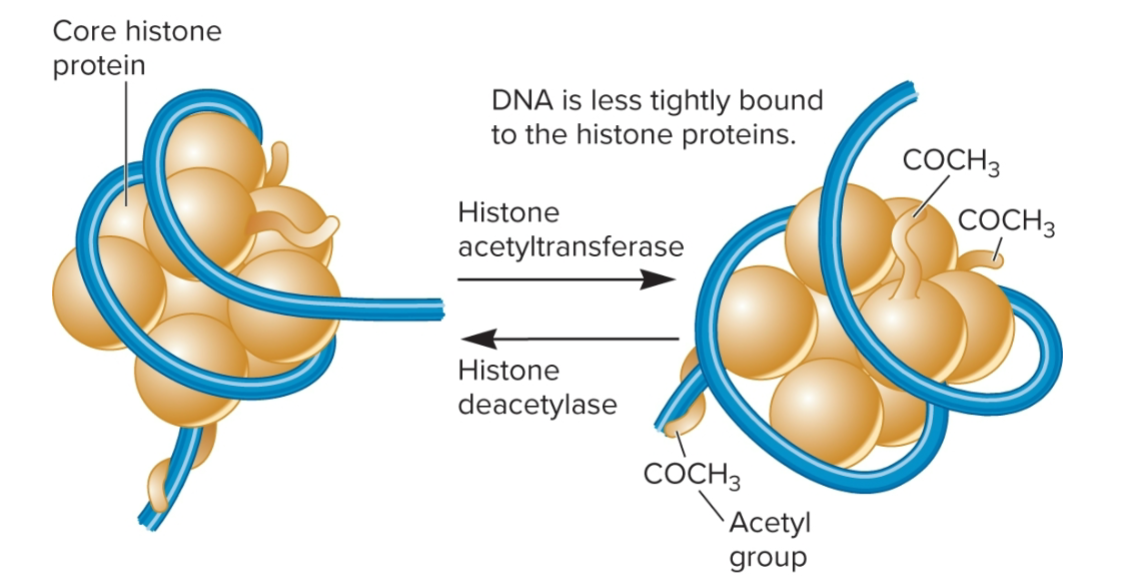
Histone deacetylases (HDACs)
remove acetyl groups from acetylated histones , favors tighter contact between histones and the DNA
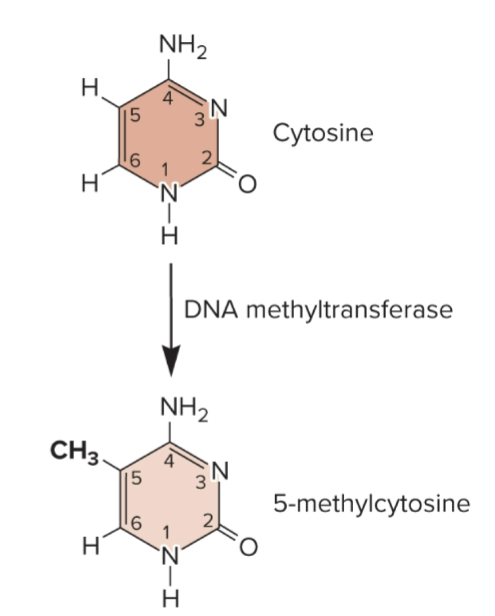
DNA methylation
covalent attachment of methyl groups (-CH3)
carried out by DNA methyltransferase
common in some euk species, but not all
DNA methylation usually inhibits euk gene transcription
yeast and Drosophila have little DNA methylation while vertebrates and plants have abundant DNA methylation
in mammals, ~ 2 to 7% of the DNA is methylated
promotes cancer by inhibiting expression of tumor-suppressor genes
CpG islands
in vertebrates and plants in many genes near promoters; 1,000-2,000 nucleotides long and contain high number of CpG sites
housekeeping genes
CpG islands are unmethylated, genes tend to be expressed in most cell types
tissue-specific gene
expression of these genes may be silenced by methylation of CpG islands
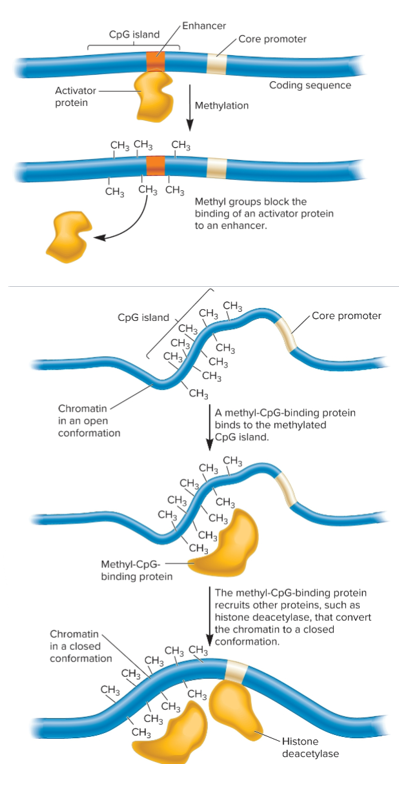
methylation influences binding of transcription factors
methyl-CpG-binding proteins recruit factors that lead to compaction of chromatin
Transcriptional silencing via methylation
Methylation inhibits binding of an activator protein
Methyl-CpG-binding protein recruits other proteins that change chromatin to closed conformation
de novo methylation
Specific genes are methylated in gametes from female or male parent, an infrequent and highly regulated process
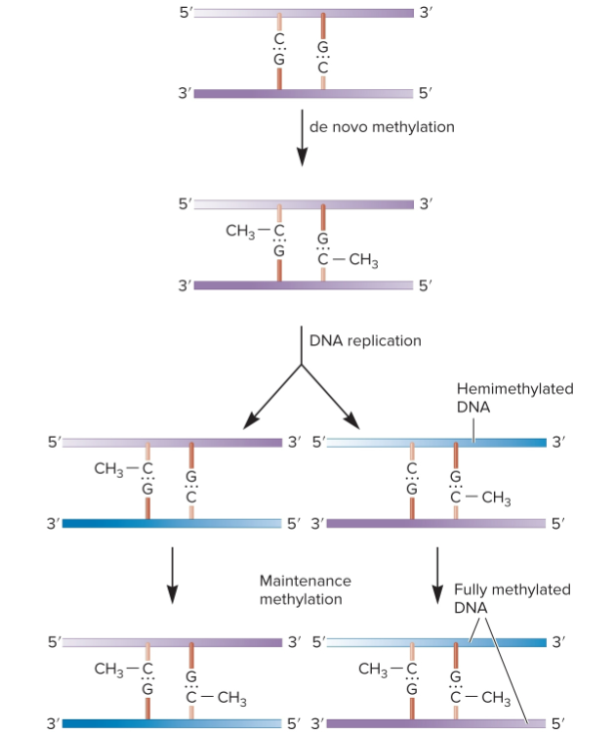
maintenance methylation
Pattern of one copy of the gene being methylated and the other is maintained in resulting offspring
How methylation is passed from mother to daughter cell
DNA, not previously methylated, becomes methylated by de novo methylation
When a fully methylated segment of DNA replicates, newly made daughter strands contain unmethylated cytosines making it hemimethylated
hemimethylated DNA recognized by DNA methyltransferase, which makes it fully methylated
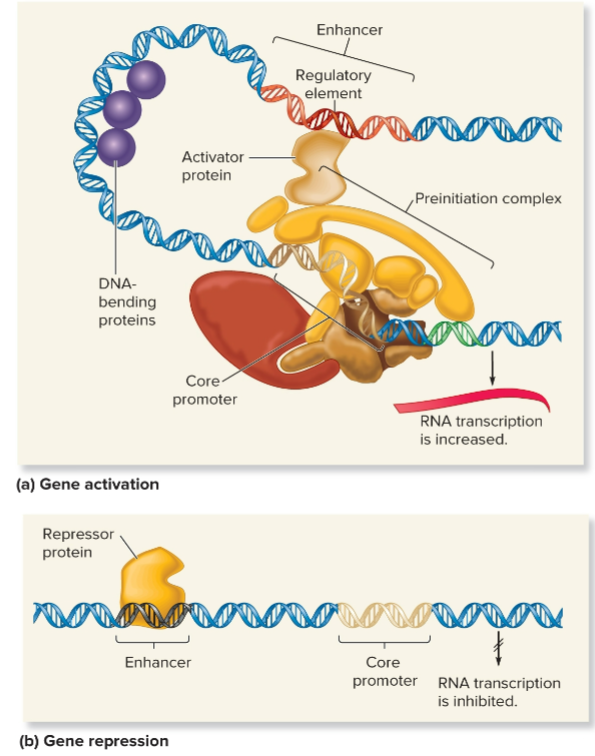
Gene activation
series of events that allow a gene to be transcribed to produce an RNA molecule
Gene activation for euks controlled by regulatory transcription factors
>1 regulatory transcription factors (activators) bind to enhancer
activators recruit coactivators (chromatin remodeling complexes and histone-modifying enzymes)
RNA pol binds to core promoter to form preinitiation complex
RNA pol proceeds to elongation phase and makes RNA transcript
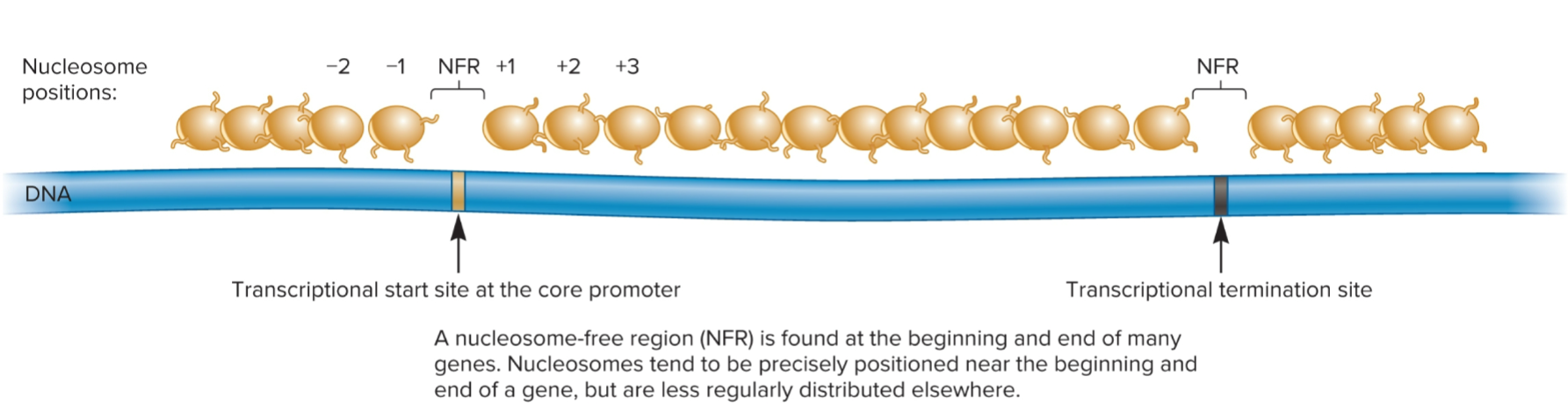
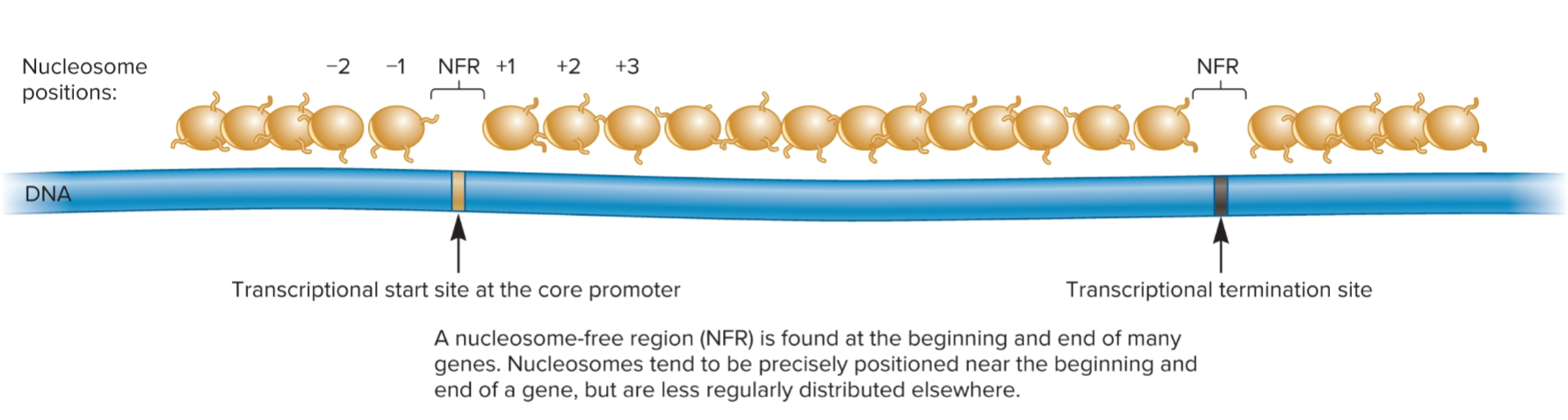
nucleosome-free region (NFR)
where transcriptional start site at core promoter is found
region of DNA where nucleosomes are not found
~150 bp in length
May be required for transcription
Not solely capable of gene activation; many genes that contain an NFR are not being actively transcribed
found at beginning and end of many genes
Nucleosomes tend to be precisely positioned near beginning and end of a gene, but are less regularly distributed elsewhere
transcriptional start site (TSS)
2 nucleosomes, −1 and +1 nucleosomes positioned on each side of NFR at transcriptional start site (TSS)
60 bp farther upstream and within NFR
+1 nucleosome contains histone variants H2A.Z and H3.3 which may also be found in −1 nucleosome and in some nucleosomes that immediately follow +1 nucleosome in transcribed region
Nucleosomes downstream from +1 nucleosome more evenly spaced near beginning of gene, but their spacing becomes less regular farther downstream
ends of many euk genes have well-positioned nucleosome followed by NFR, important for transcriptional termination
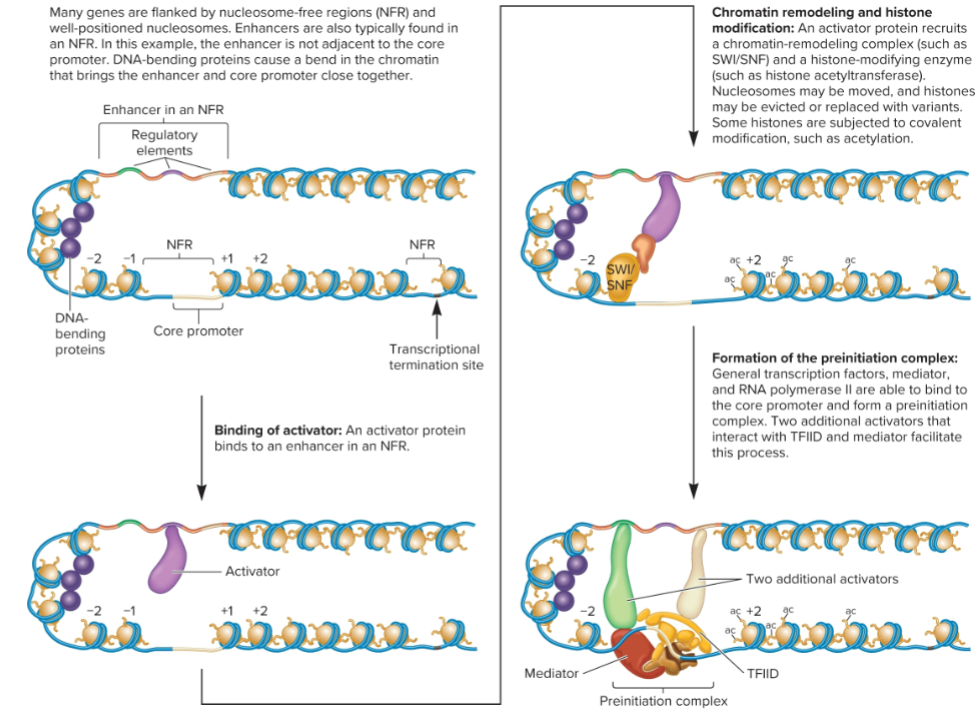
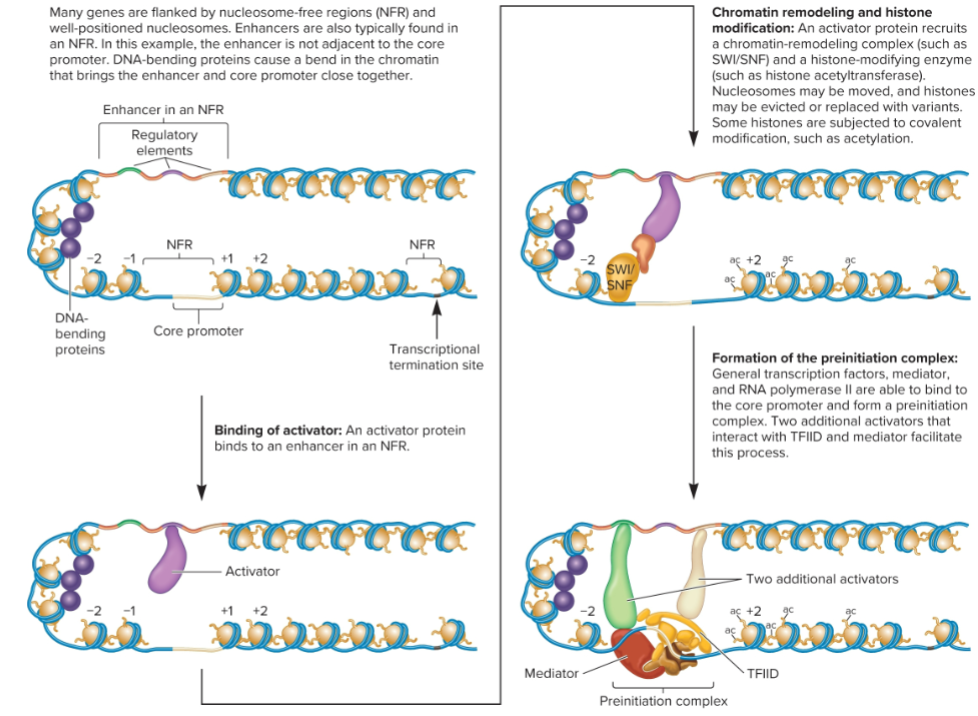
Transcriptional activation
involves changes in nucleosome position and composition and modifications to histone
Activators, may bind within NFR near core promoter or at distance enhancers, recruit chromatin remodeling complexes and histone-modifying enzymes to promoter
Binding of activators (1/4 Changes that occur to form a Pre-initiation Complex in transcription)
Activation protein binds directly to DNA of regulatory elements within enhancer sequences. enhancer can be close or far from transcriptional start site
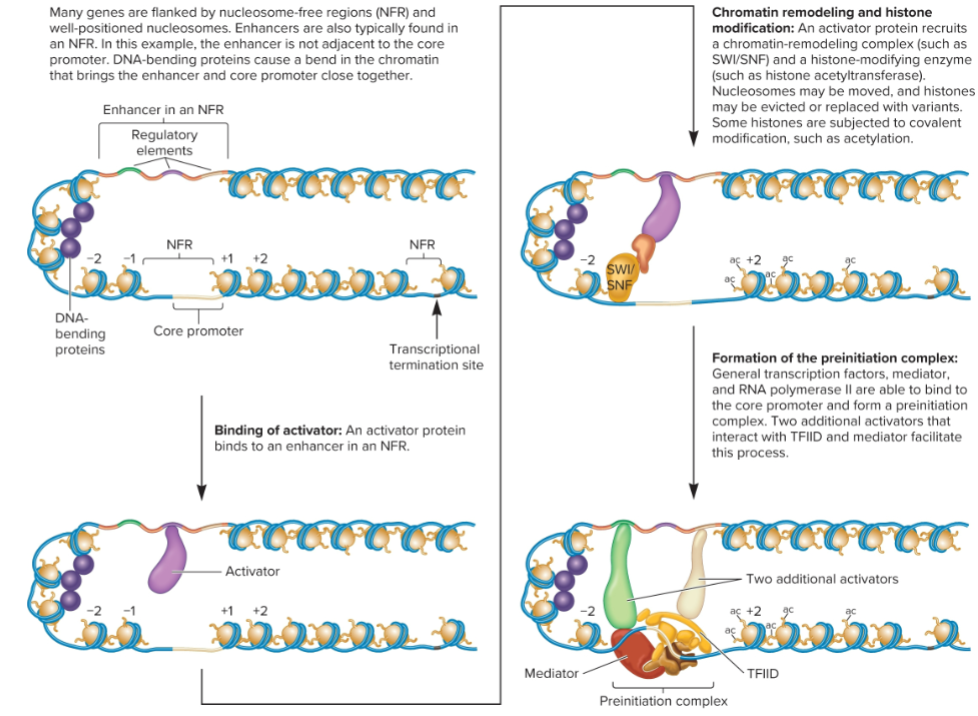
Recruitment of coactivators (2/4 Changes that occur to form a Pre-initiation Complex in transcription)
Activators recruit coactivators
increase transcription rate but don’t bind directly to DNA
can enhance transcription (chromatin remodeling, histone modification, recruitment or stimulation of the preinitiation complex, and stimulation of transcriptional elongation)
Chromatin remodeling and histone modification (3/4 Changes that occur to form a Pre-initiation Complex in transcription)
activator protein recruits chromatin remodeling complex and histone-modification enzyme
nucleosome may be moved, and histone may be evicted or replaced with variants
some histones are subjected to covalent modification.
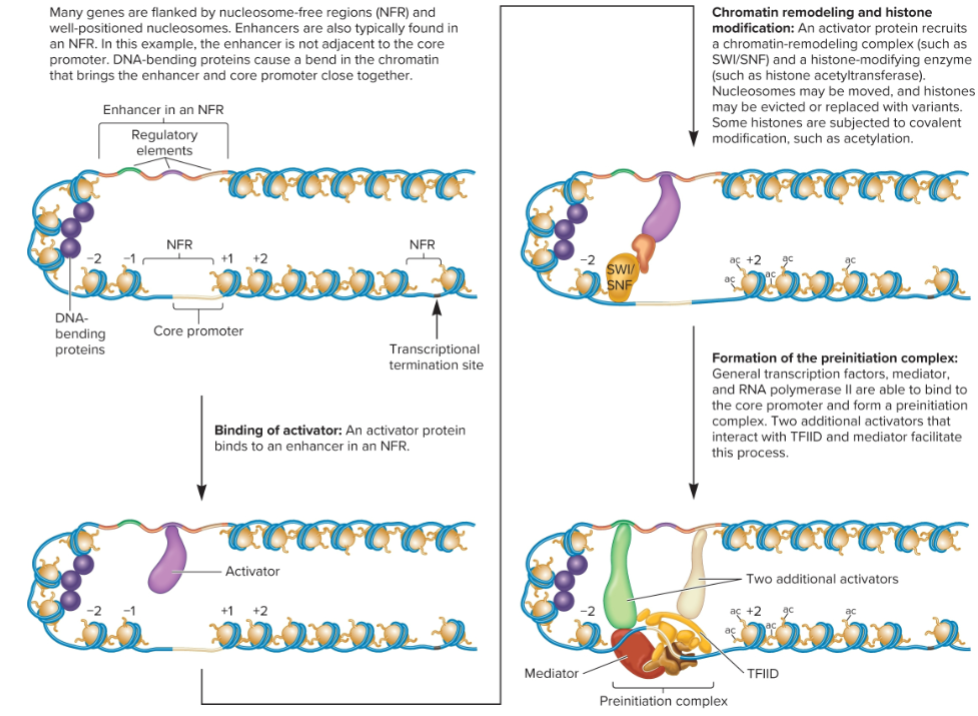
Formation of preinitiation complex (4/4 Changes that occur to form a Pre-initiation Complex in transcription)
General transcription factors and RNA pol II bind to core promoter and form preinitiation complex.
Formation of open complex (1/4 events during elongation in transcription)
DNA strands must be separated into open complex so that one can act as template for RNA synthesis
TFIIH has subunit that functions as DNA translocase, separating DNA strands to convert the closed complex to an open complex
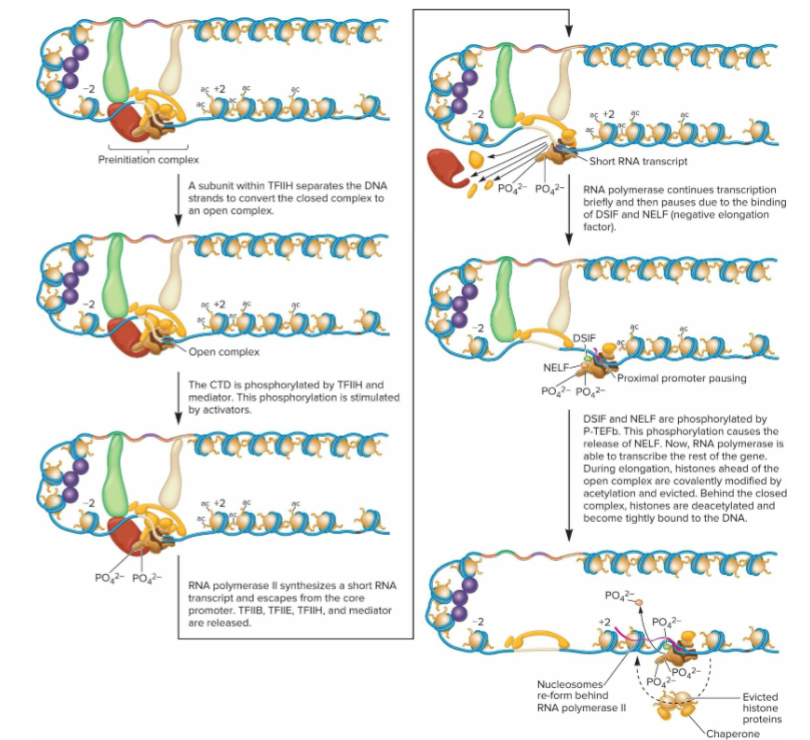
Promoter escape (2/4 events during elongation in transcription)
At core promoter, GTFs and mediator bind to RNA pol II and prevent it from traveling along template strand
For elongation to occur, promoter escape must happen (RNA pol II released from this binding)
Phosphorylation of CTD allows promoter escape.
Transcriptional activators facilitate switch between initiation and elongation stages.
Proximal promoter pausing (3/4 events during elongation in transcription)
regulates euk genes
RNA pol II pauses in RNA synthesis while close to transcriptional start site
Involves binding of 2 factors; DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF)
to release pause, positive transcriptional elongation factor b (P-TEFb) phosphorylates both DSIF and NELF
results in release of NELF and causes DSIF to facilitate elongation
RNA pol II can now transcribe rest of gene
Other proposed roles of pausing during elongation in transcription
Regulating level of transcription
Helping maintain nucleosome-free region by blocking nucleosome assembly over core promoter
Providing time for recruitment of factors in RNA modifications
Providing time for binding of transcriptional elongation factors that provide stability and facilitate transcription process
Histone modifications (4/4 events during elongation in transcription)
Histone-modifying enzymes important in histone removal and replacement through histone acetylation, H3 methylation, and H2B ubiquitination
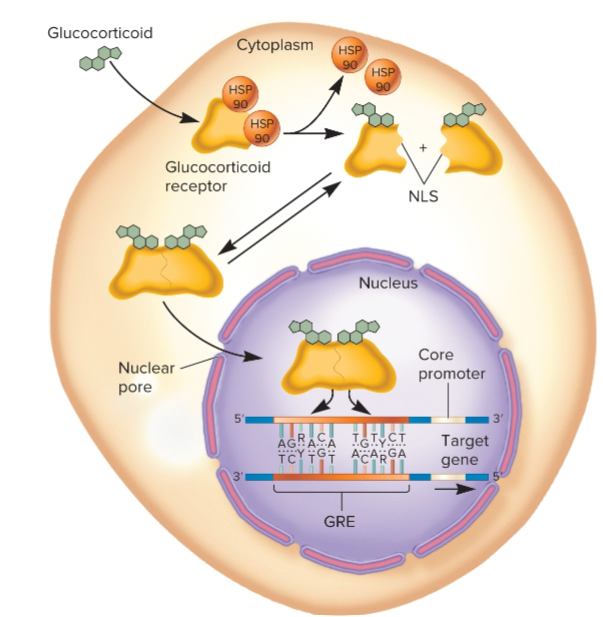
steroid receptors
Regulatory transcription factors that respond to steroid hormones
hormone binds to transcription factor to affect gene transcription
Steroid hormones produced by endocrine glands and secreted into bloodstream before being taken up by cells that respond to hormone
Glucocorticoids
influence nutrient metabolism in most cells, promote glucose utilization, fat mobilization and protein breakdown
GRE (Glucocorticoid Response Elements)
found within enhancers
located near dozens of different genes, so hormone can activate many genes
consists of 2 sequences that are close together
5’-AGRACA-3’
3’-TCYTGT-5’
glucocorticoid hormones enter cell and bind to glucocorticoid receptor subunits that dimerize, enter nucleus, bind to GRE, and activate gene transcription
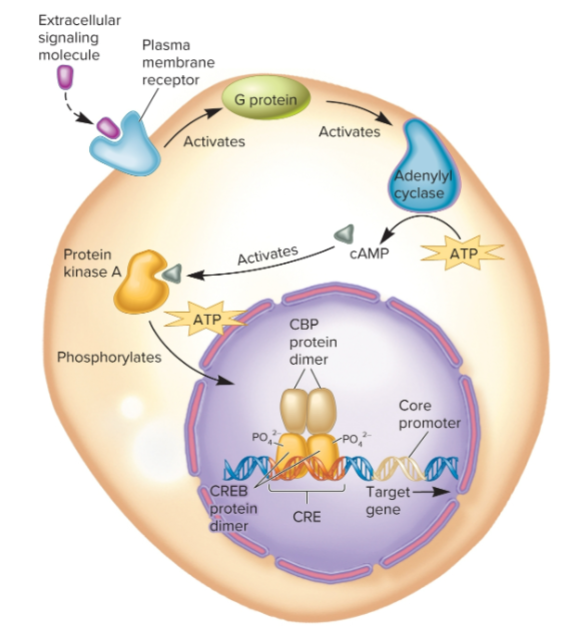
CREB (cAMP response element-binding) protein
regulatory transcription factor that responds to cAMP which acts as a second messenger by activating protein kinase A, which phosphorylates CREB, allowing it to bind to coactivator CBP
binding of CBP causes transcription of adjacent gene to be greatly increased
Unphosphorylated CREB can bind to DNA, but cannot activate RNA pol
becomes activated in response to extracellular cell-signaling molecules that cause an increase in cytoplasmic concentration of cAMP
binds to 2 adjacent sites with consensus sequence
5’-TGACGTCA-3’
3’-ACTGCAGT-5’
a CRE (cAMP response element)
Gene repression
any mechanism that inhibits transcription of a gene, resulting in a lower level of RNA synthesis from that gene
can be short-term, by directly inhibiting steps needed for gene activation or long-term, as in gene silencing via formation of heterochromatin
Repressor
protein that binds directly to a DNA sequence, such as a regulatory element within an enhancer, and inhibits transcription
exert their effects by interacting with other proteins or protein complexes called corepressors
Transcriptional Regulation in Bacteria, Archaea, and Eukaryotes
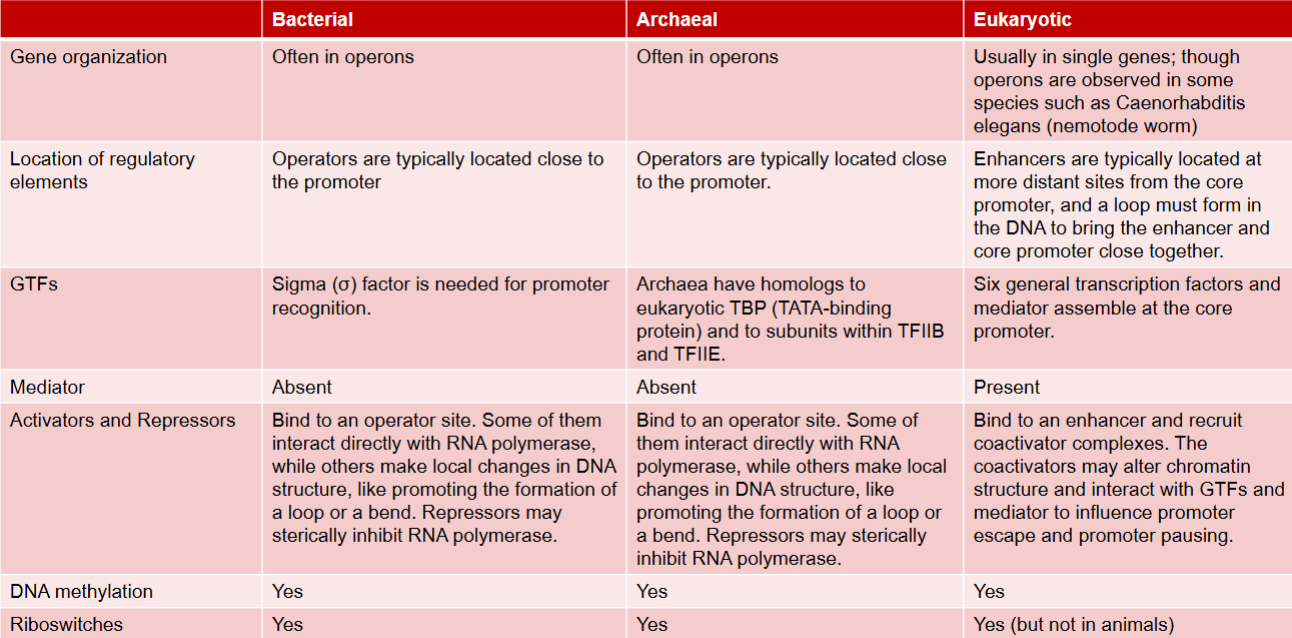
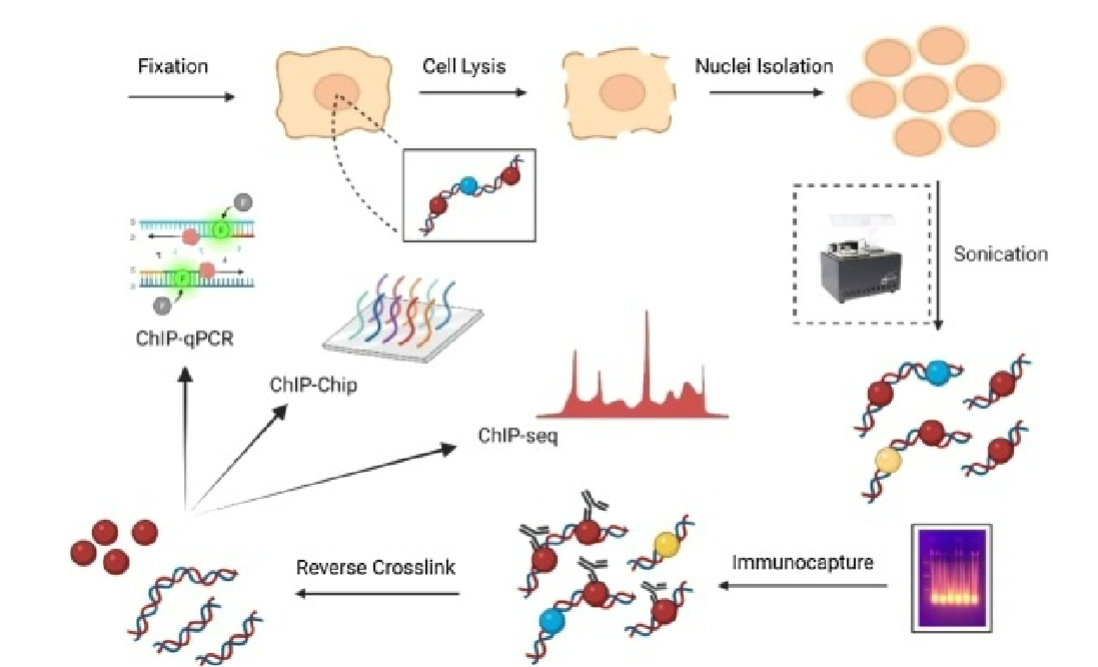
Chromatin immunoprecipitation (ChIP)
technique used to study DNA- protein interactions in cells
involves crosslinking proteins to associated DNA, fragmenting DNA, immunoprecipitating protein-DNA complex using antibody, and then analyzing purified DNA
allows identification of which proteins are bound to specific DNA regions and can provide insights into gene regulation and epigenetic processes
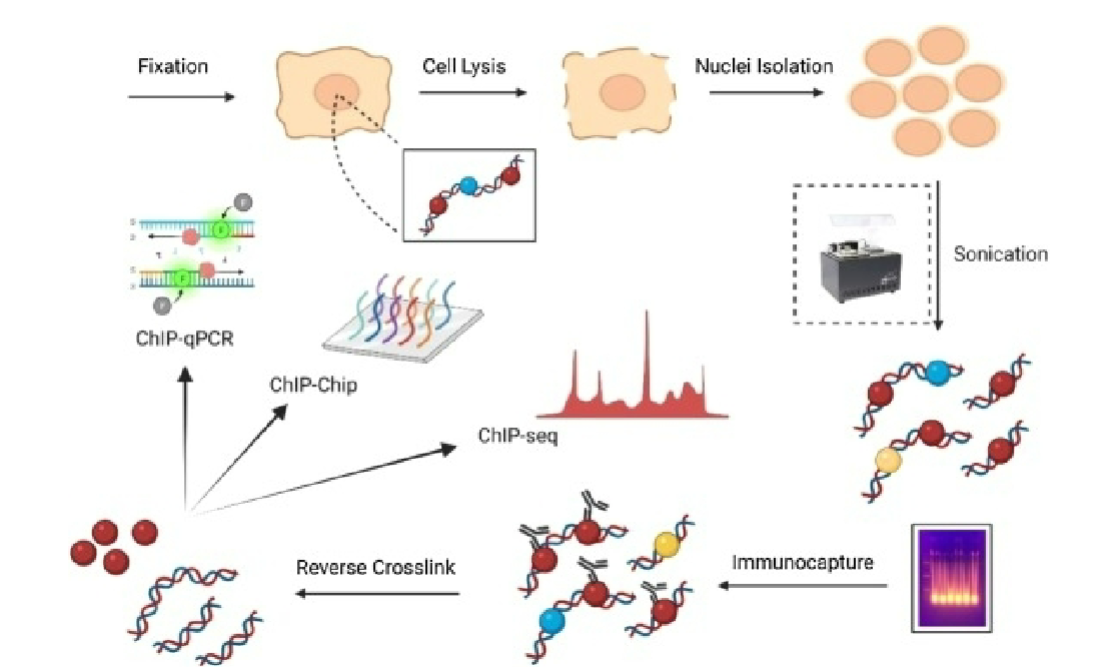
Crosslinking (ChIP)
DNA and proteins are crosslinked in live cells using formaldehyde or UV light (or other crosslinkers), stabilizing their interactions
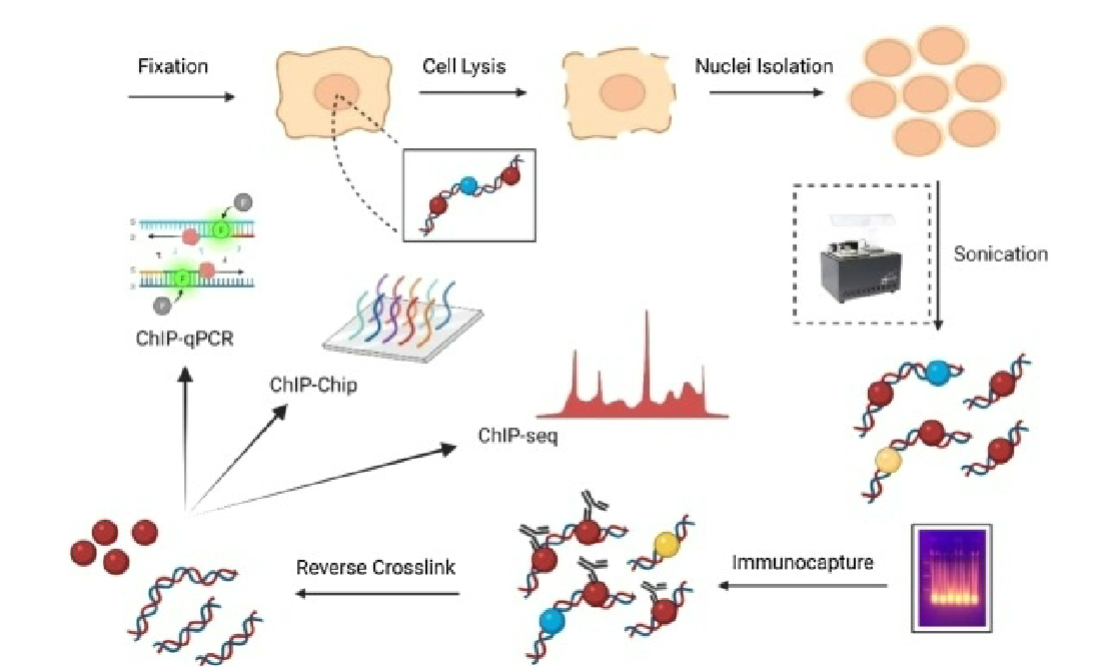
Chromatin Fragmentation (ChIP)
crosslinked chromatin is fragmented using sonication or enzymatic digestion to create smaller DNA fragments
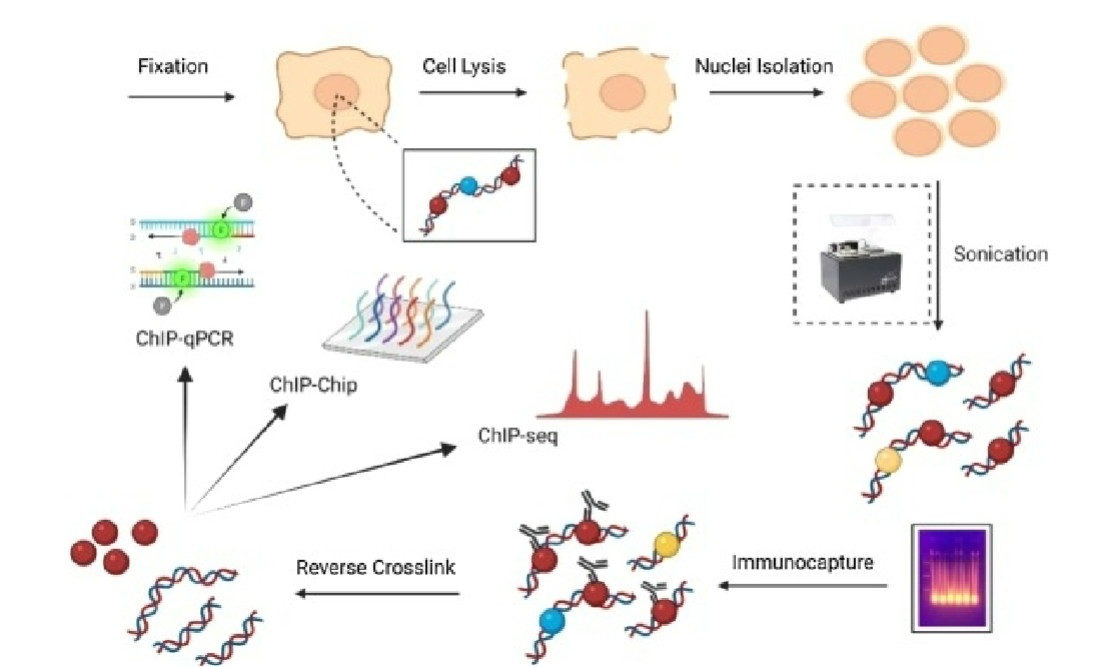
Immunoprecipitation (ChIP)
antibody specific for protein of interest is used to capture DNA-protein complex from fragmented chromatin
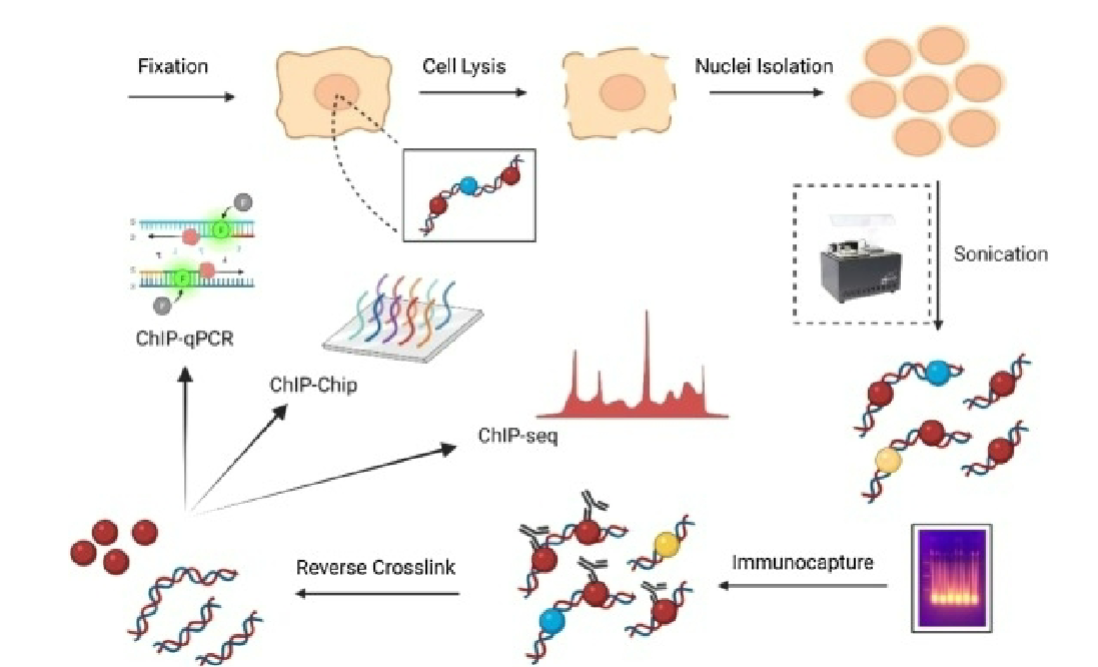
DNA Purification and Analysis (ChIP)
DNA associated with precipitated protein-DNA complex is purified and analyzed, can involve PCR, microarrays, or ChIP-sequencing (ChIP-Seq) to identify specific DNA regions where protein is bound.
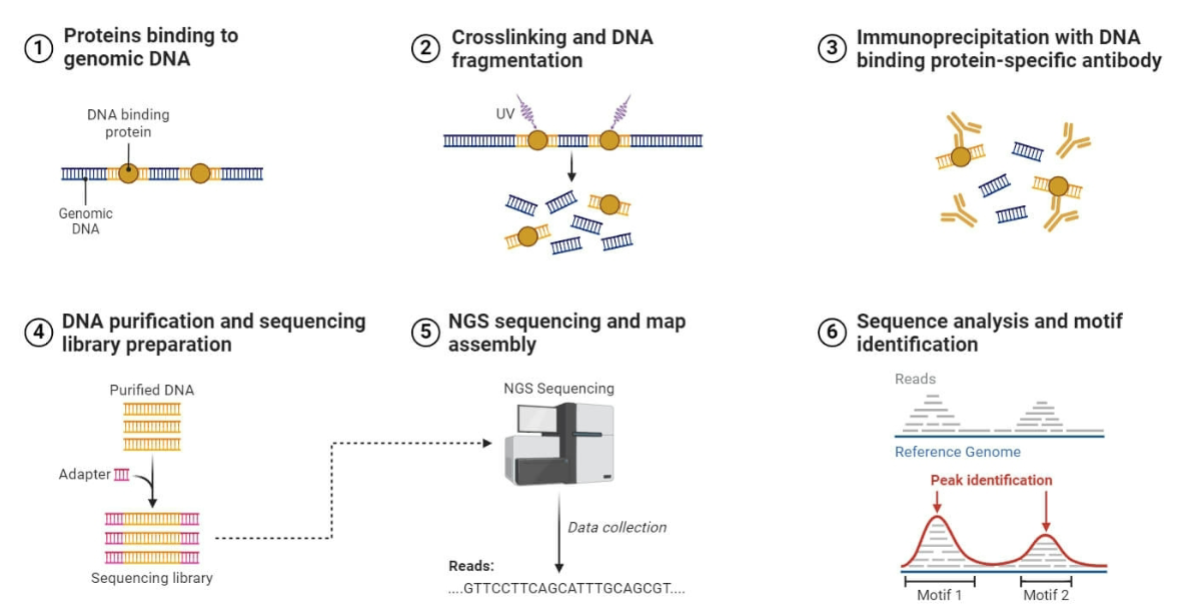
ChiP-Seq
Maps locations of specific nucleosomes within a genome
Allows determination of the location of nucleosomes, histone variants and where covalent modifications of histones occur
Chromatin immunoprecipitation combined with DNA sequencing
Performed in species where genome has been sequenced
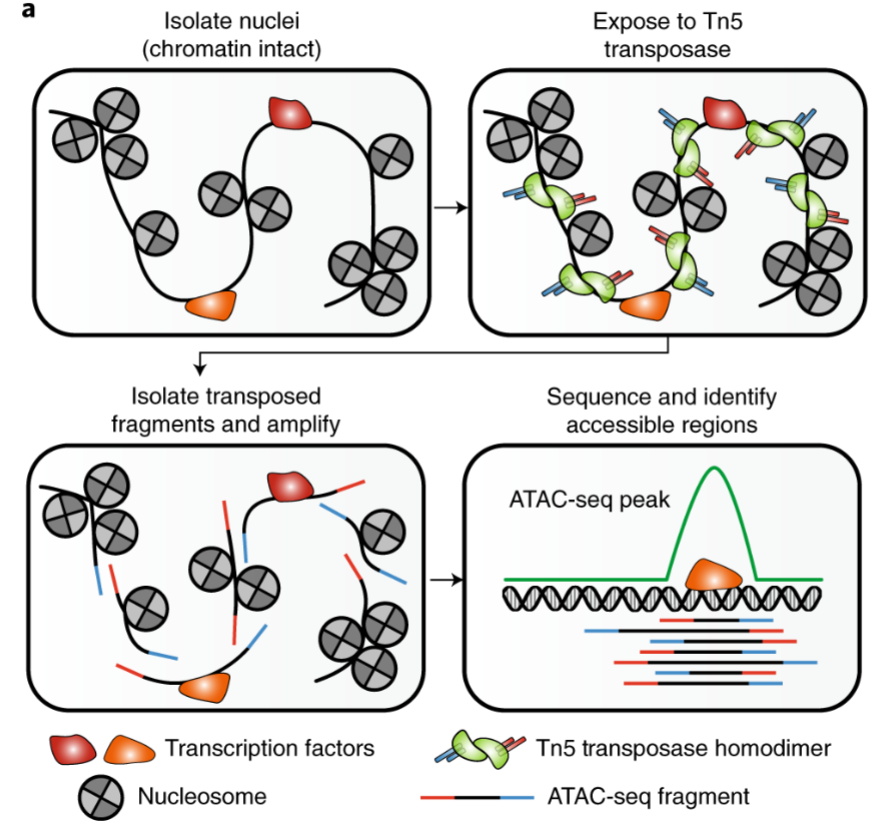
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing)
high-throughput sequencing method used to assess genome-wide chromatin accessibility
provides insight into regulatory landscape of genome by identifying regions where DNA is accessible for binding by cellular factors
uses hyperactive Tn5 transposase to tag accessible chromatin regions, allowing for identification of open chromatin regions and their interplay with other factors.
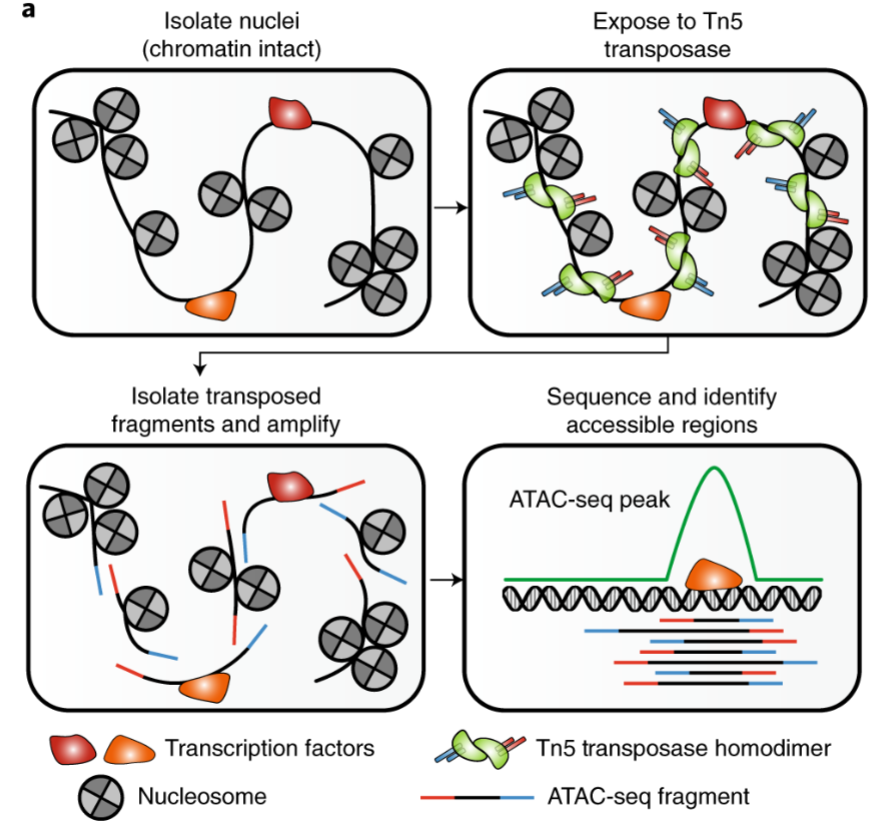
4 steps of ATAC-seq
Cell Lysis and Nuclei Preparation: cell nuclei isolation and gentle lysis to maintain chromatin integrity
Tagmentation: Tn5 transposase used to insert sequencing adapters into open chromatin regions, simultaneously tags and fragments DNA
Library Preparation and Sequencing: tagged and fragmented DNA is then purified, amplified, and sequenced using next-generation sequencing
Data Analysis: sequencing reads are aligned to reference genome, and peak calling algorithms used to identify regions of increased chromatin accessibility
Epigenetics
mechanisms that lead to changes in gene expression that can be passed and are reversible, but do not involve change in sequence of DNA
epimutation: heritable change in gene expression that does not alter the sequence of DNA
Epigenetic inheritance: epigenetic changes passed from parent to offspring
not all epigenetic changes are passed from parent to offspring
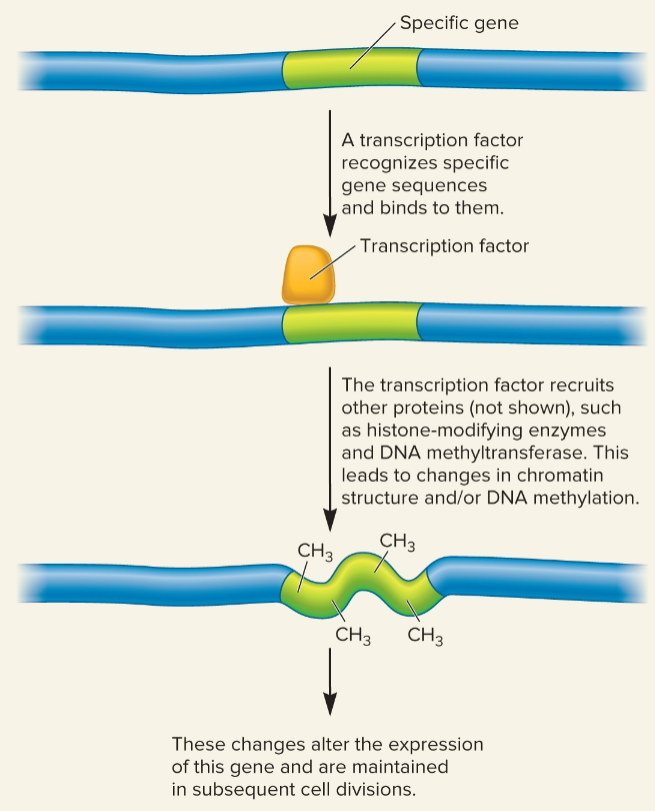
Targeting a gene for epigenetic modification by a transcription factor
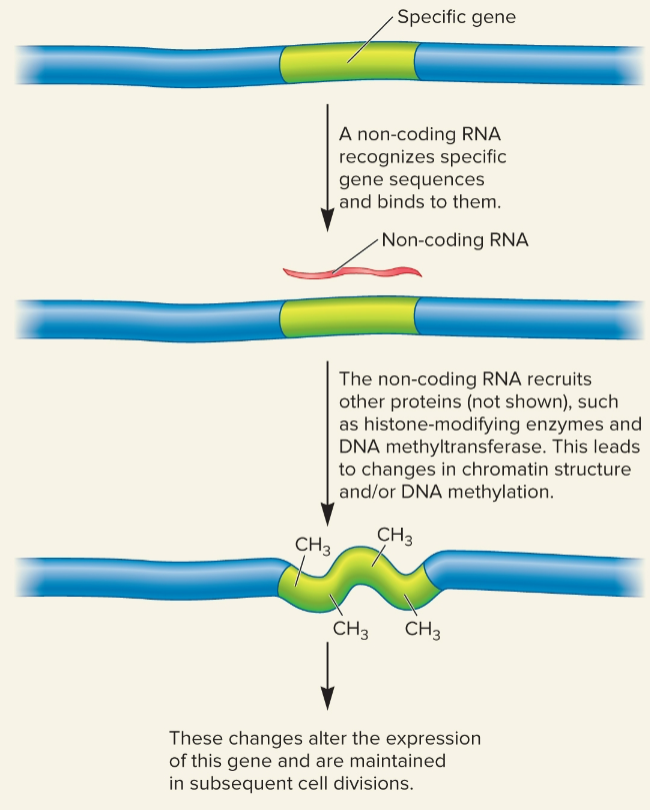
Targeting a gene for epigenetic modification by a noncoding RNA
cis-epigenetic changes
maintained at a specific site
affect only one copy of gene but not other copy
maintained during cell division
in subsequent cell divisions, methylated copy of gene B is always methylated whereas other remains unmethylated
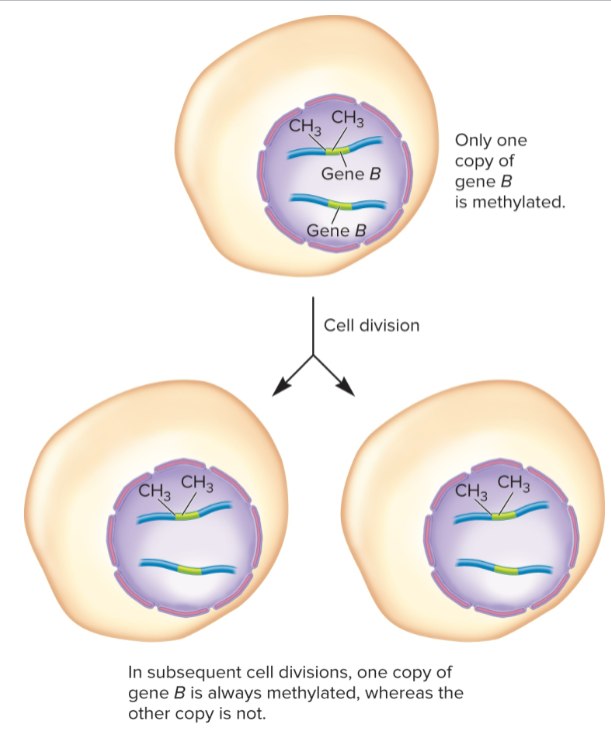
trans-epigenetic changes
maintained by diffusible factors, such as transcription factors, affects both copies of a gene
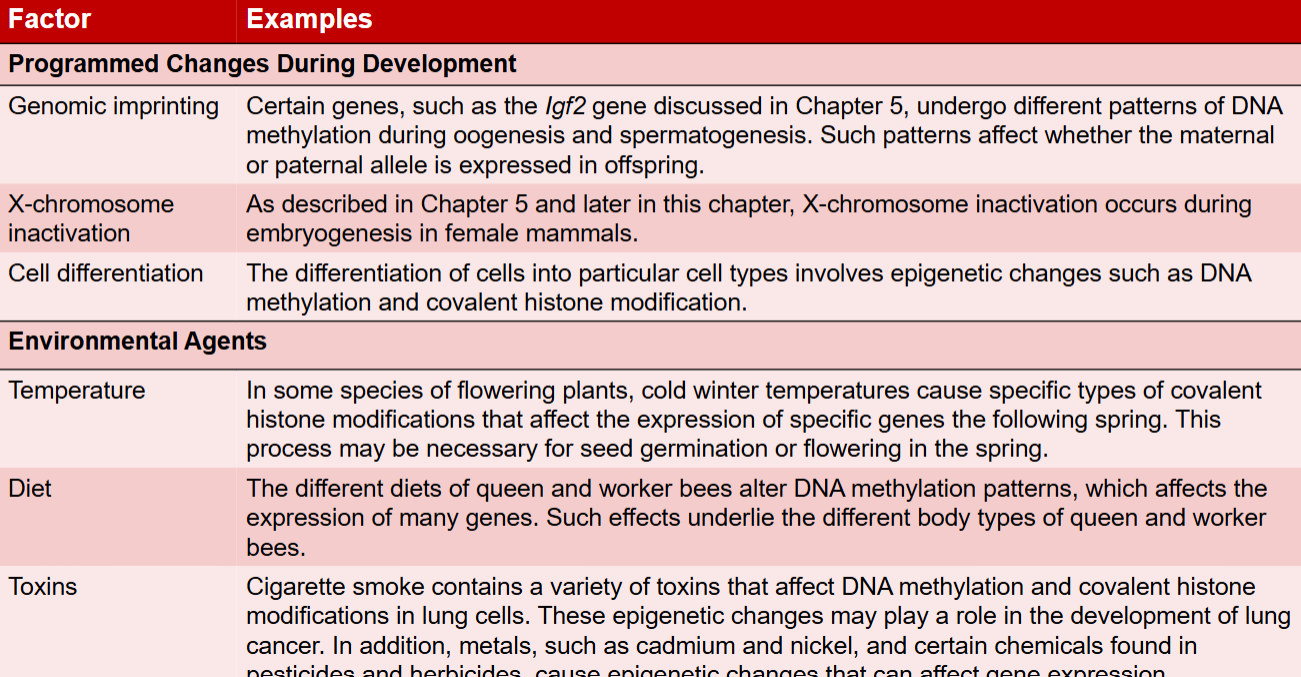
Factors that promote epigenetic changes
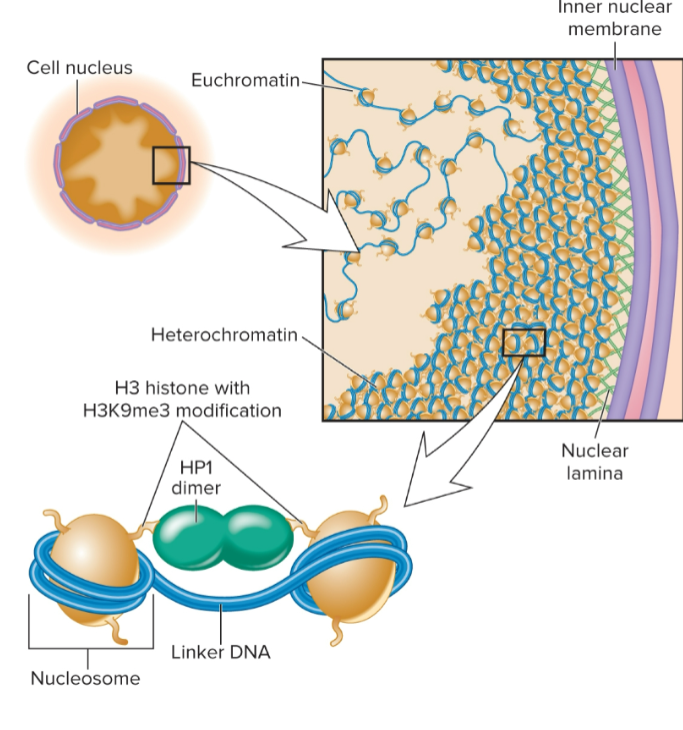
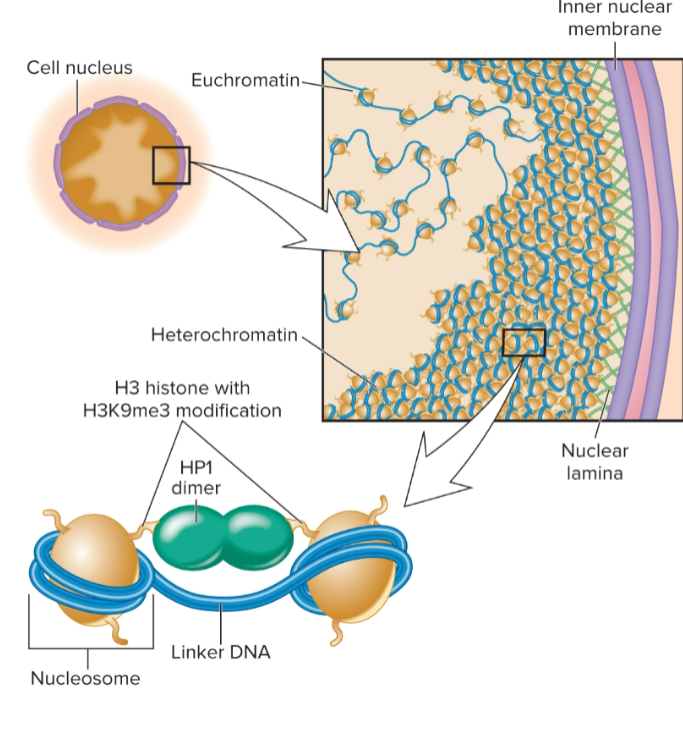
general features of eukaryotic chromatin structure
Nucleosomes are basic unit
146 bp of DNA wrapped around octamer of histone proteins (H2A, H2B, H3, and H4)
Nucleosomes interact in a zigzag manner
Loop domains formed from SMC proteins and CTCFs
Chromatin composed of DNA, protein, non-coding RNAs
Euchromatin
regions that are not stained during interphase
transcriptionally active
occupies a central position in nucleus
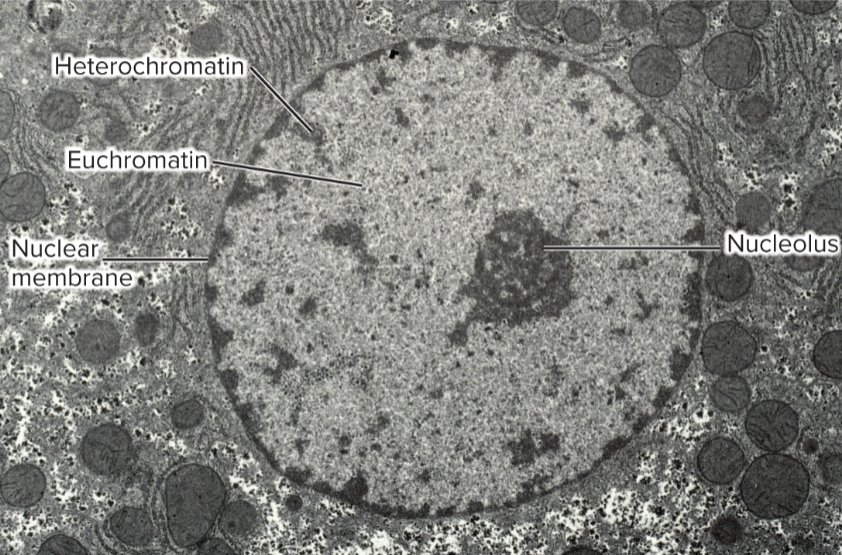
Heterochromatin
regions that are stained throughout cell cycle
greater level of compaction
localized along periphery of nucleus; attached to nuclear lamina
Gene silencing: inhibition of transcription; may limit access of activators or other aspects of transcription
Prevention of transposable element movement: genes needed for transposition are silenced
Prevention of viral proliferation: genes needed to produce more viruses are silenced
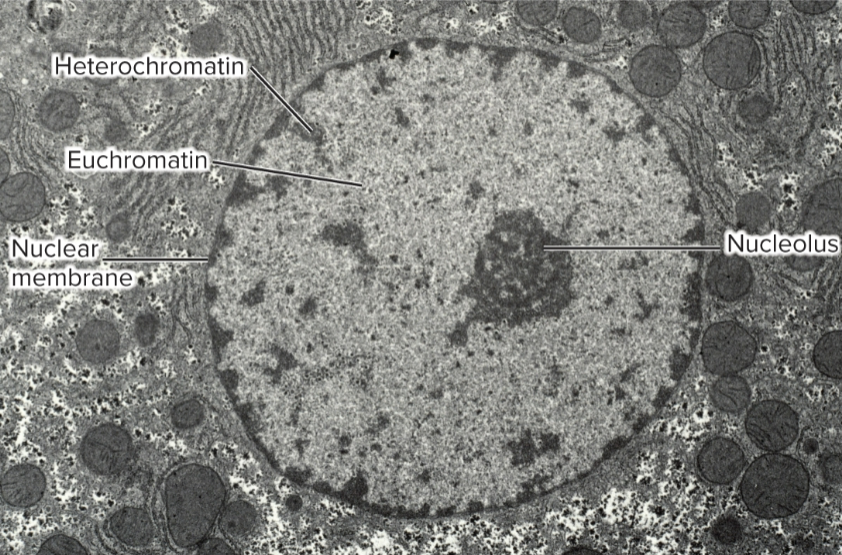
Constitutive heterochromatin
heterochromatic at same location in all cell types
chromosomal location: close to centromere or telomere
repeat sequences: many short tandemly repeated sequences
DNA methylation: highly methylated on cytosines in vertebrates and plants
Histone modifications: H3K9me3 common in constitutive heterochromatin in yeast and animals; H3K9me2 in plants
Facultative heterochromatin
locations vary among different cell types
allows silencing of genes in a cell specific manner
formation is reversible; depends on stage of development or cell type
chromosomal location: multiple sites between centromere and telomere
repeat sequences: LINE-type repeats
DNA methylation: methylation at CpG islands in gene regulatory regions; silences genes
histone modifications: H3K9me3 found in facultative heterochromatin; animals have H3K27me
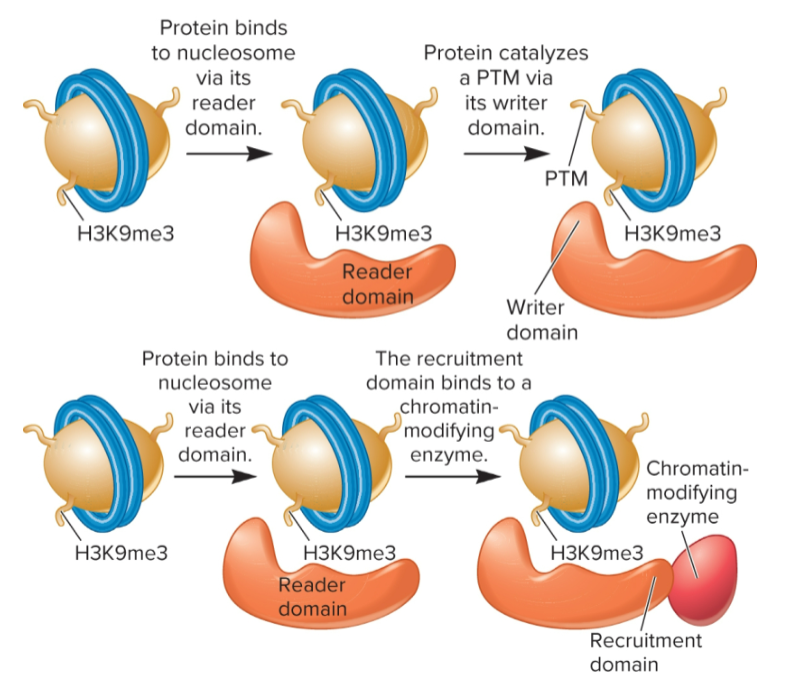
posttranslational modifications (PTMs)
on amino-terminal tails of histone proteins
specific proteins bind to particular PTMs in nucleosomes via protein domains
reader, writer, eraser, and/or recruitment domains
writer vs eraser vs recruitment domains
writer domains: addition of PTMs
eraser domains: remove PTMs
recruitment domains: recruit other proteins, such as chromatin remodelers or chromatin-modifying enzymes
Molecular events leading to heterochromatin with Higher Order Structure
histone PTMs
binding of proteins to nucleosomes
chromatin remodeling
DNA methylation
binding of non-coding RNAs
higher-order (reproducible in 3D) structural features in heterochromatin
has closer, more stable contacts of nucleosomes with each other via HP1 which recognizes H3K9me3 and bridges nucleosomes; makes them more compact
forms closer loop domains, SMCs promote loop domain formation, CTCFs form a crosslink that stabilizes loops
binds to the nuclear lamina
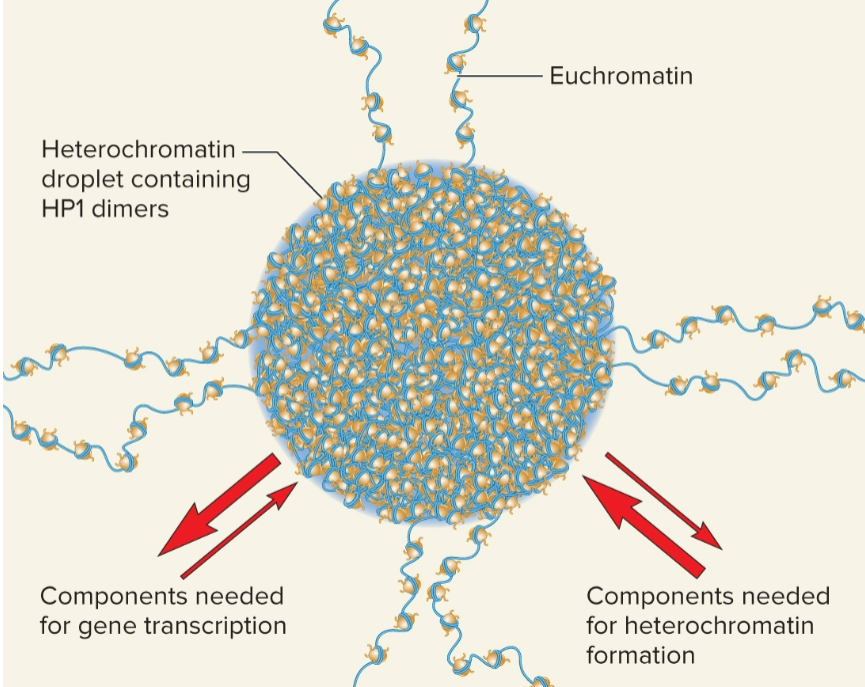
may undergo liquid-liquid phase separation
Lamina-associated domains (LADS)
chromosomal regions associated with nuclear lamina (NL); fibrous layer of proteins
organize chromosomes into chromatin territories
involved in gene repression
Liquid-liquid phase separation (LLPS)
formation of liquid-like compartments formed by macromolecules that become concentrated in a given location and come out of solution
nucleolus, located inside nucleus, is formed by LLPS
Heterochromatin may undergo LLPS
HP1 protein
forms a dimer that binds to two nucleosomes carrying H3K9me3 modification, holds two nucleosomes in close association
3 stages of formation of facultative and constitutive chromatin
Nucleation: short chromosomal site bound by chromatin-modifying enzymes and chromatin-remodeling complexes
Spreading: adjacent euchromatin is turned into heterochromatin
Barrier: in interphase chromosomes, spreading stops when it reaches a barrier
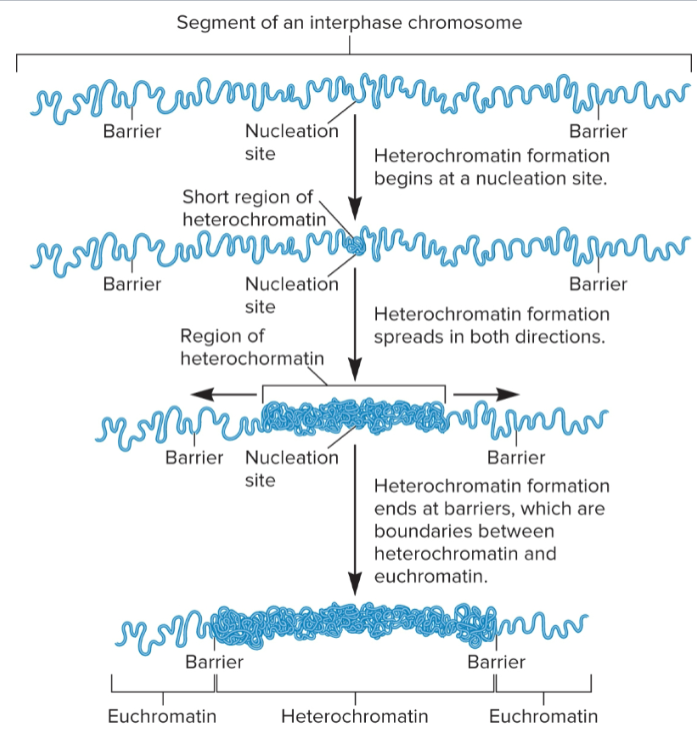
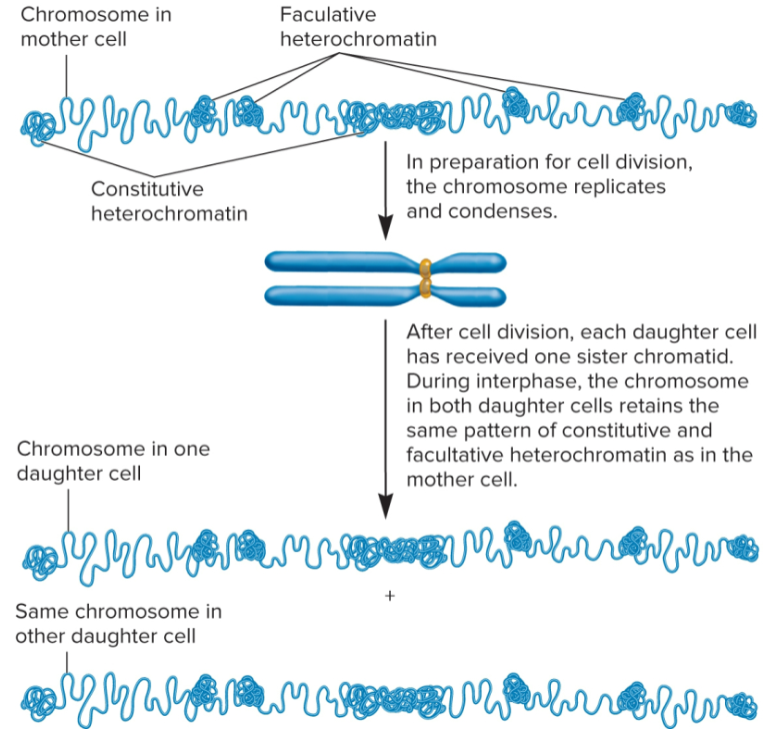
Pattern of heterochromatin before and after cell division
pattern of constitutive and facultative heterochromatin is same in daughter cells as it was in mother cell
Multicellular species: heterochromatin patterns established during embryonic development
Constitutive: same in all cell types
Facultative: cell specific
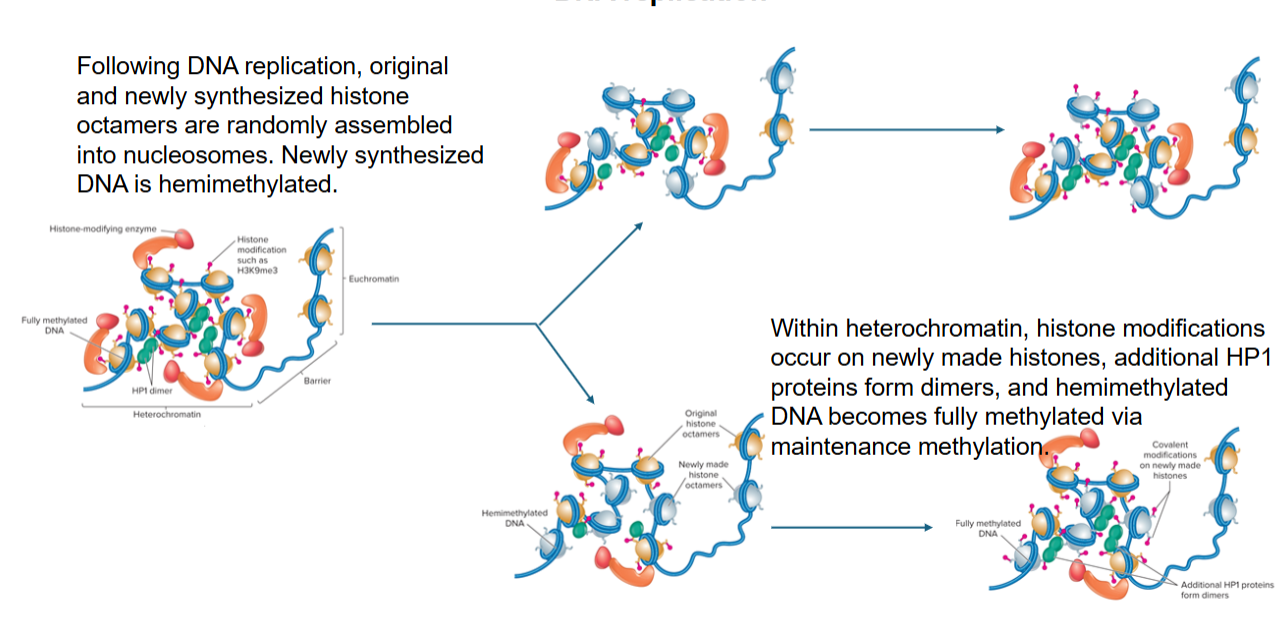
During and following cell division, heterochromatin structure is maintained by
DNA methylation: hemimethylated DNA becomes fully methylated via maintenance methylation
Histone modifications: histones recruit chromatin-modifying enzymes and chromatin-remodeling complexes to daughter chromatids
DNA pol: recruit chromatin-modifying complexes
Local chromatin structure: higher-order structure favors reformation of heterochromatin
ICF syndrome
immunodeficiency, centromere instability, and facial anomalies, can be due to a mutation in a DNA methyltransferase gene
Roberts syndrome
prenatal growth defects, craniofacial abnormalities, limb malformations, mutations in a gene for an acetyltransferase
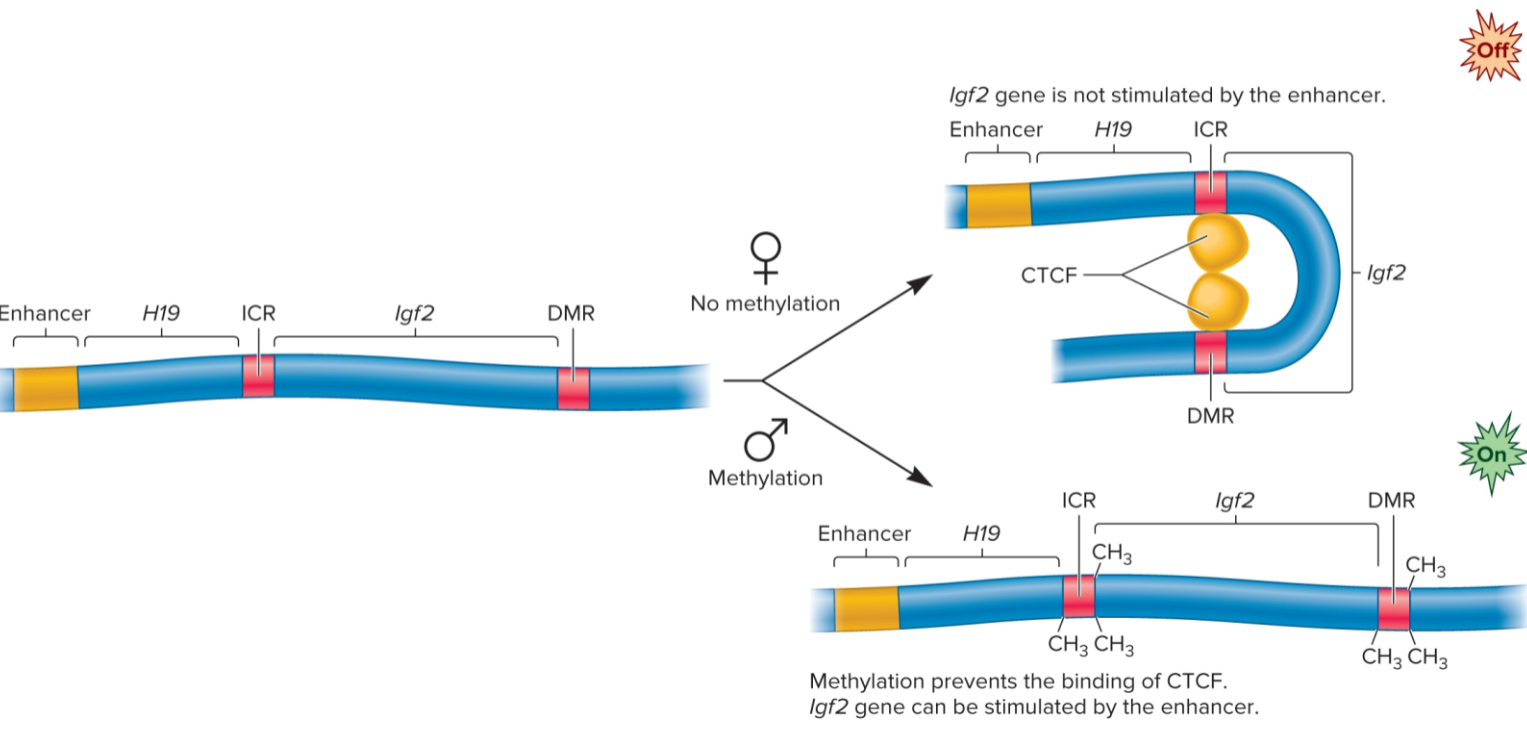
Genomic imprinting
form of gene regulation in which offspring expresses copy of gene from one parent but not both
in mammals, only Igf2 gene inherited from father is expressed
methylation inhibits binding of CTC-binding factor, which allows Igf2 gene to be stimulated by a nearly enhancer
CTC-binding factor binds to unmethylated gene and inhibits transcription by stabilizing loop
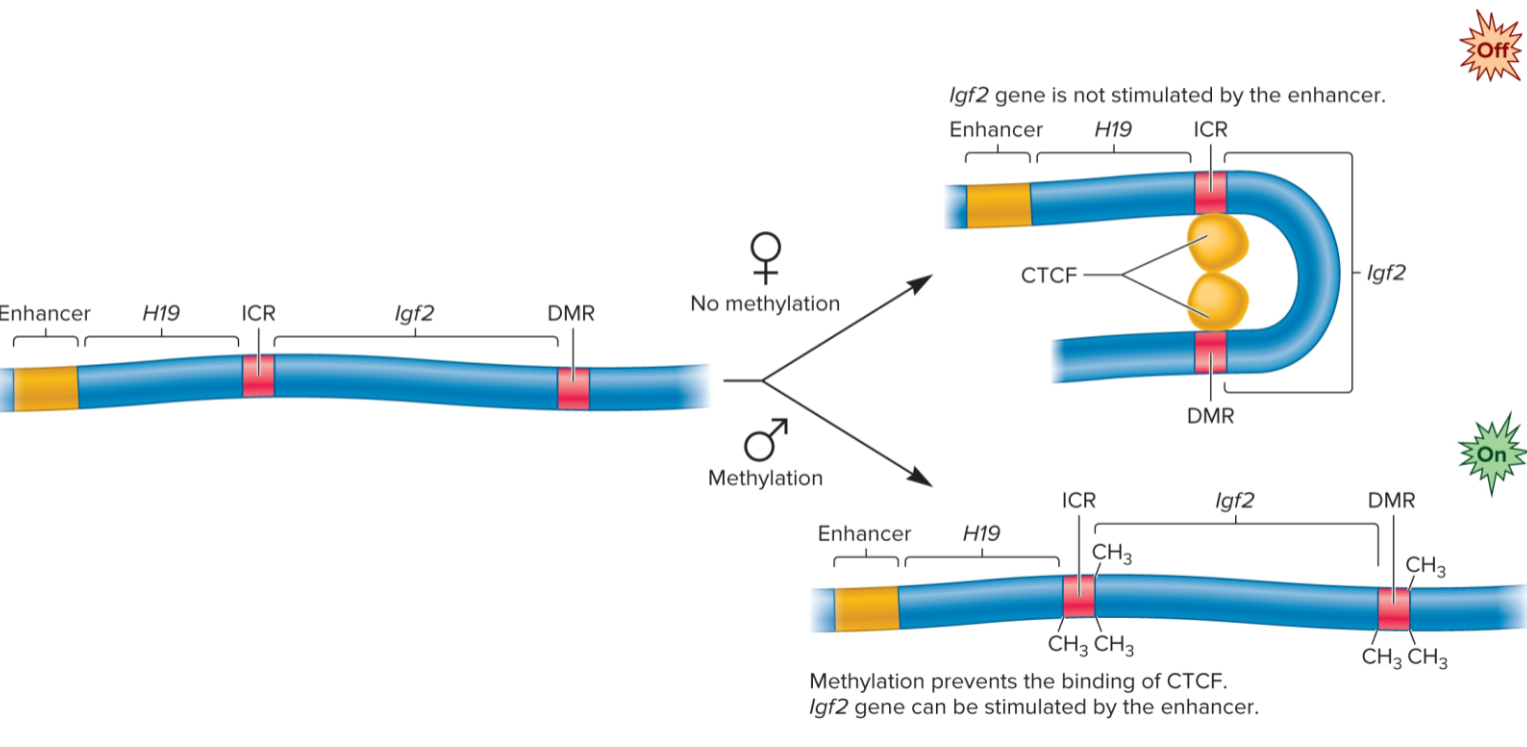
Igf2 gene (genomic imprinting)
gene is de novo methylated during sperm formation but not during egg formation
methylation occurs at two sites: imprinting control region (ICR) and a differentially methylated region (DMR)
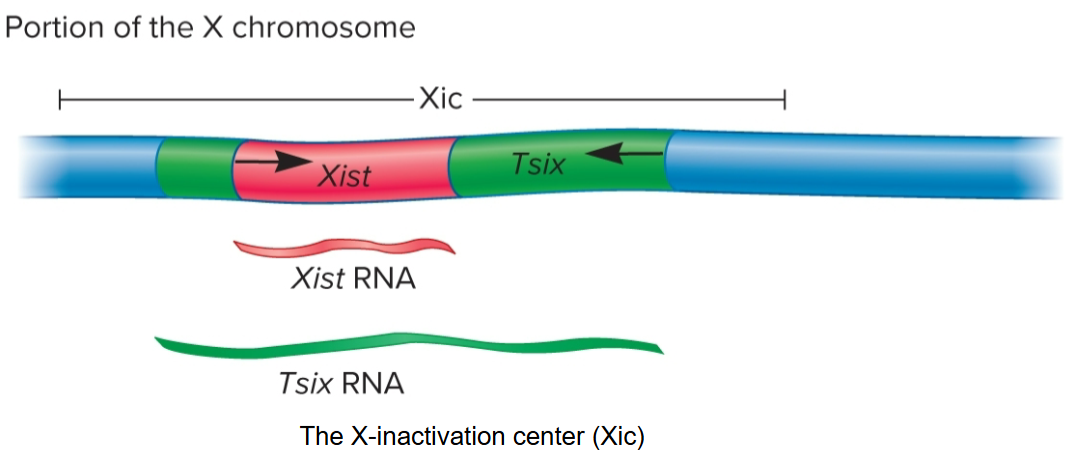
X-chromosome inactivation (XCI)
occurs during embryogenesis in female mammals
X-inactivation center (Xic) found on X chromosome
Xic contains two genes, Xist and Tsix, which are transcribed in opposite directions
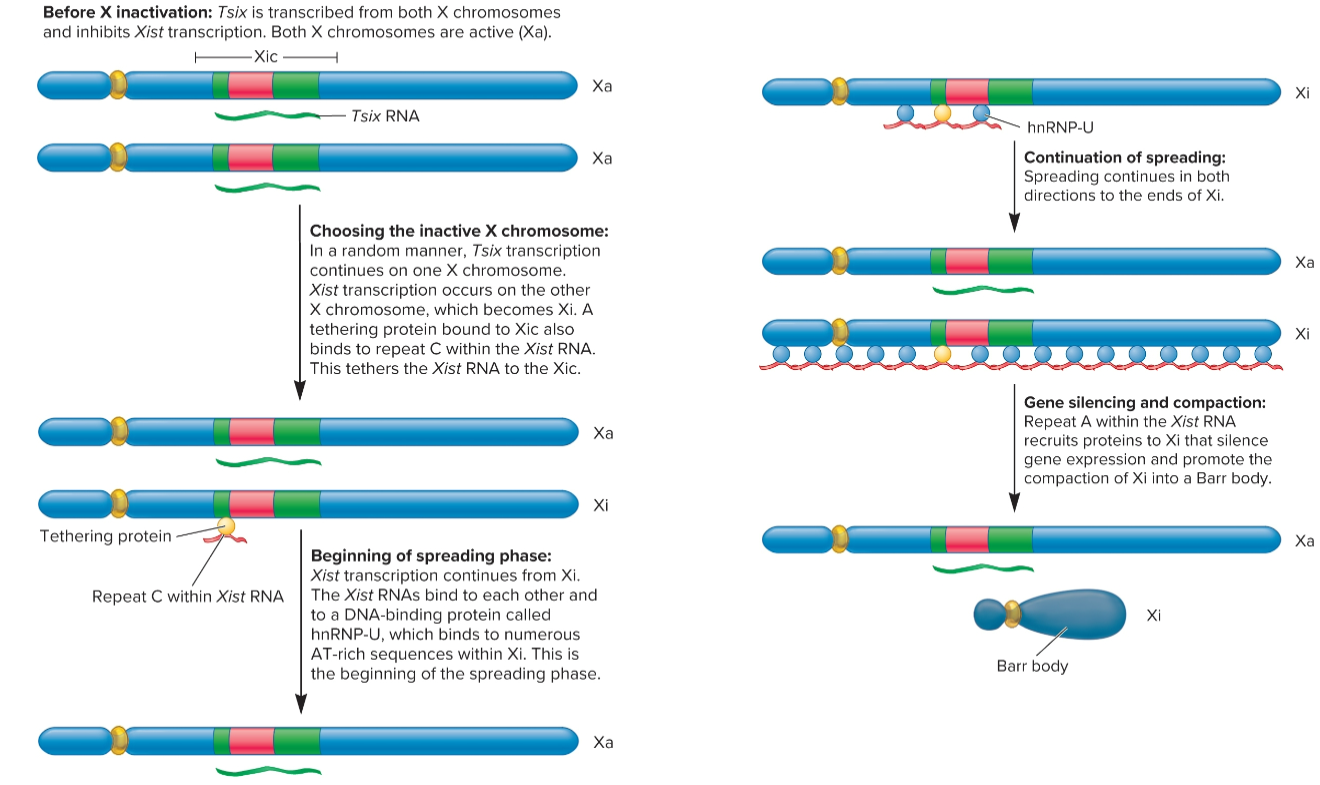
process of X-chromosome inactivation
one of X chromsomes (the one that will become inactive Barr body) begins to express Xist gene, process of choosing inactive X chromosome is not well understood
Xist RNA binds to XIC and spreads to both ends of X chromosome
Xist RNA recruits proteins to X chromosome that make it into a compact Barr body, inactive with regard to gene expression
some genes on this chromosomes may be expressed
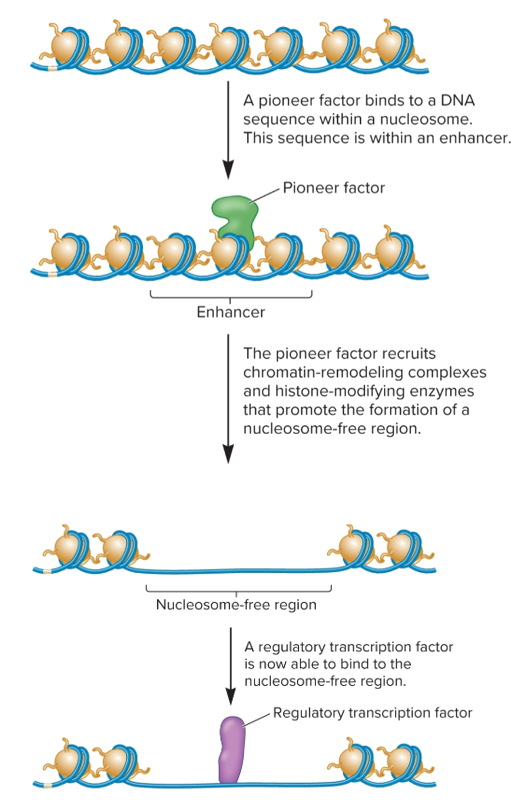
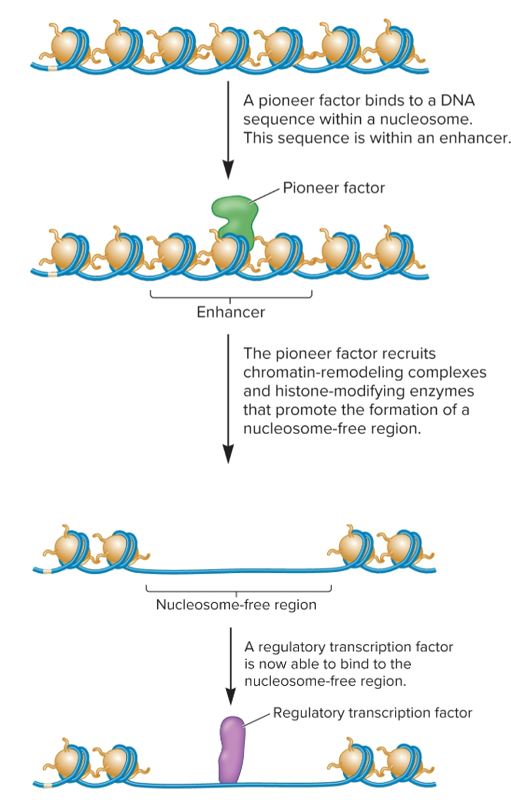
Pioneer factors
transcription factors that can recognize and bind to DNA sequences exposed on surface of a nucleosome
recruit chromatin-remodeling complexes and histone-modifying enzymes that carry out epigenetic changes (histone eviction and covalent modifications)
influence ability of other transcription factors to bind to enhancer sequences
can decrease level of DNA methylation by binding to CpG islands, blocking access by DNA methyltransferases
involved in activation/silencing of some genes
Pioneer factors during embryonic development
Play a role in changing chromatin structure, has positive/negative effects on transcription
Drive reprogramming of genome during initial steps of development
Enable some genes to be activated/repressed
Work in conjunction with nonpioneer transcription factors to promote cell differentiation
Prime certain genes for later expression
Important in differentiated cells in adults
Levels of expression vary during different stages of embryonic development and among different cell types
Trithorax group (TrxG) vs Polycomb group (PcG)
key regulators of epigenetic changes during development that produce specific cell types and tissues
Trithorax group (TrxG): involved with gene activation
Polycomb group (PcG): involved with gene repression
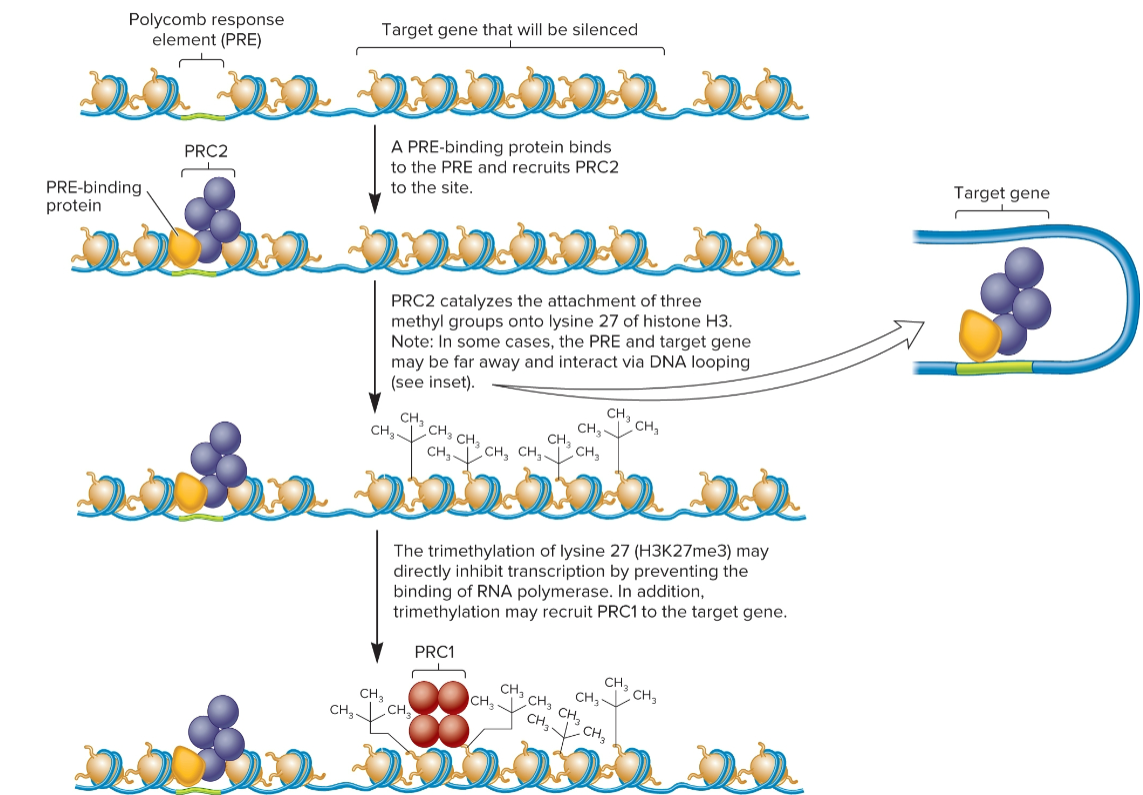
polycomb group complex
PRC1 and PRC2
repression begins w/ binding of PRC2 to polycomb response element (PRE)
leads to trimethylation of lysine 27 on histone H3.
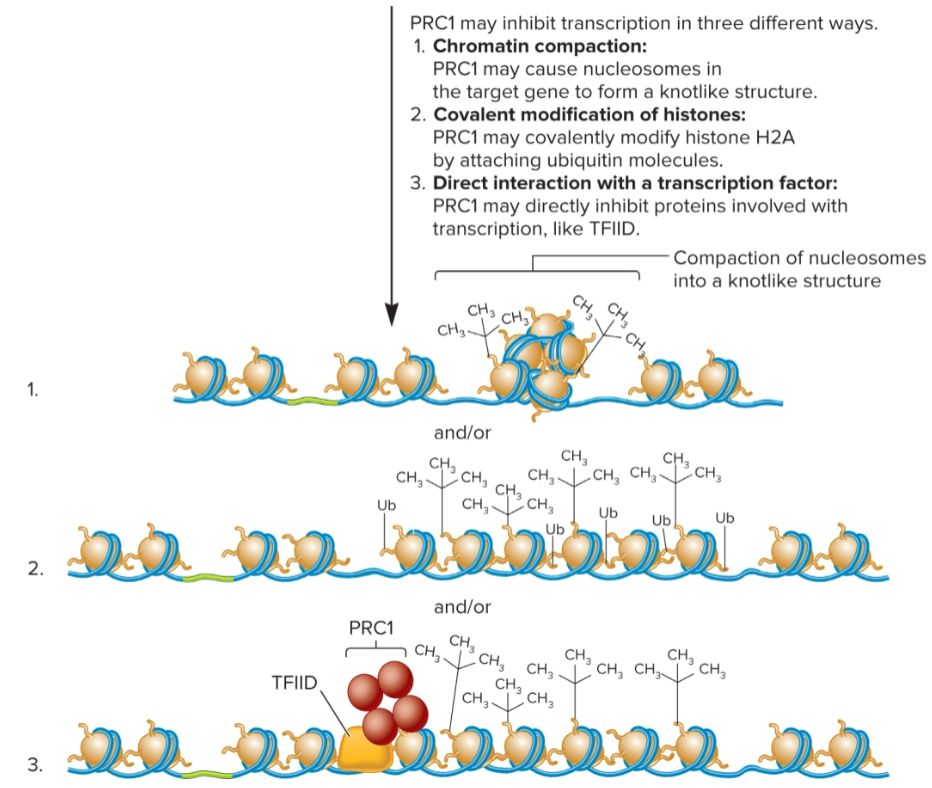
3 ways PRC1 inhibits transcription
Chromatin compaction: PRC1 causes nucleosomes in target gene to form a knot-like structure
Covalent modification of histones: PRC1 covalently modifies histone H2A by attaching ubiquitin molecules
Direct interaction with transcription factor: PRC1 directly inhibits proteins involved with transcription, like TFIID.
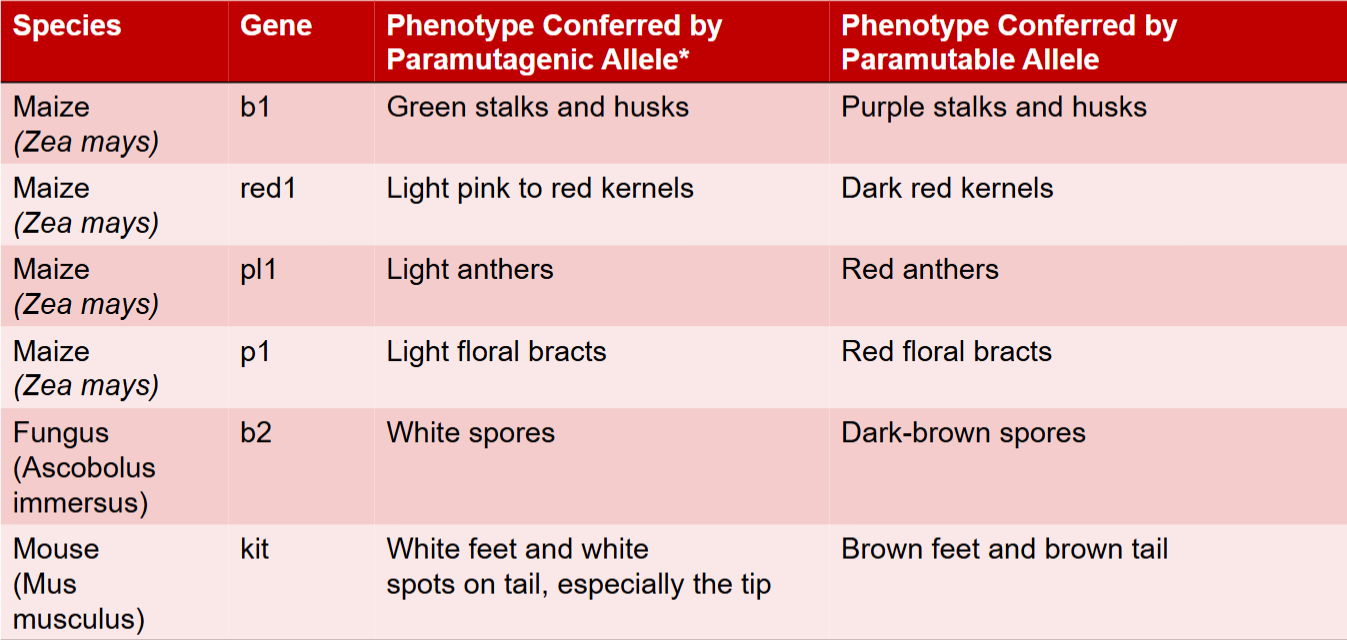
paramutagenic vs paramutable
Interaction between two alleles at single locus where one allele induces a change in other allele
Allele that has this capacity is paramutagenic
Allele that has been altered is paramutable
Example: b1 locus in maize
paramutations
weaker expression of a paramutagenic allele is due to epigenetic changes that decrease/silence its transcription
epigenetic changes transferred to paramutable allele
silencing of paramutagenic alleles occurs using short ncRNA molecules
multiple tandem repeat sequences located close to the coding sequences of paramutagenic and paramutable alleles and may be used to make siRNAs
functional mop1 gene (mediator of paramutation 1 gene) and RNA-dependent RNA pol is required for paramutation
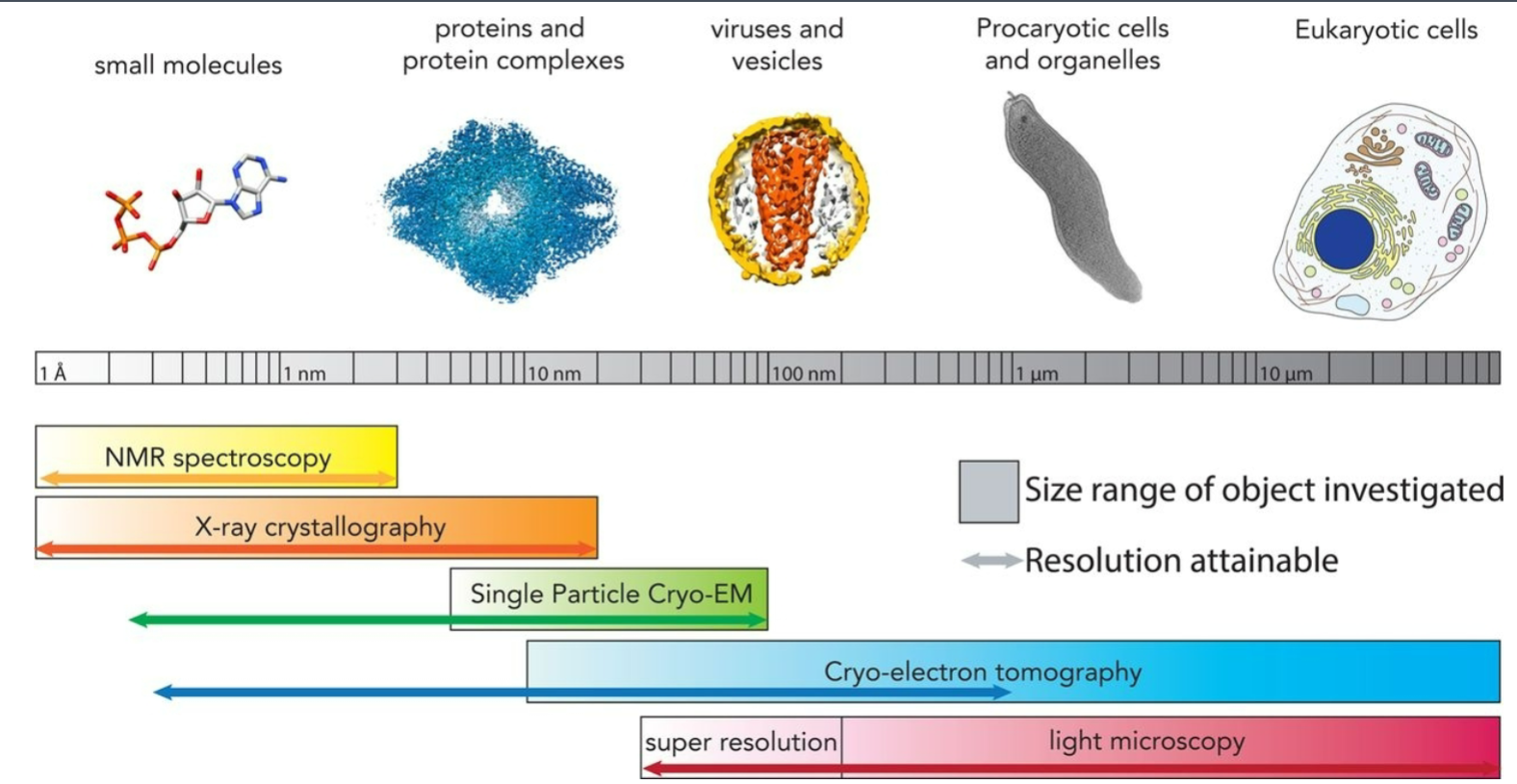
microscopy and size
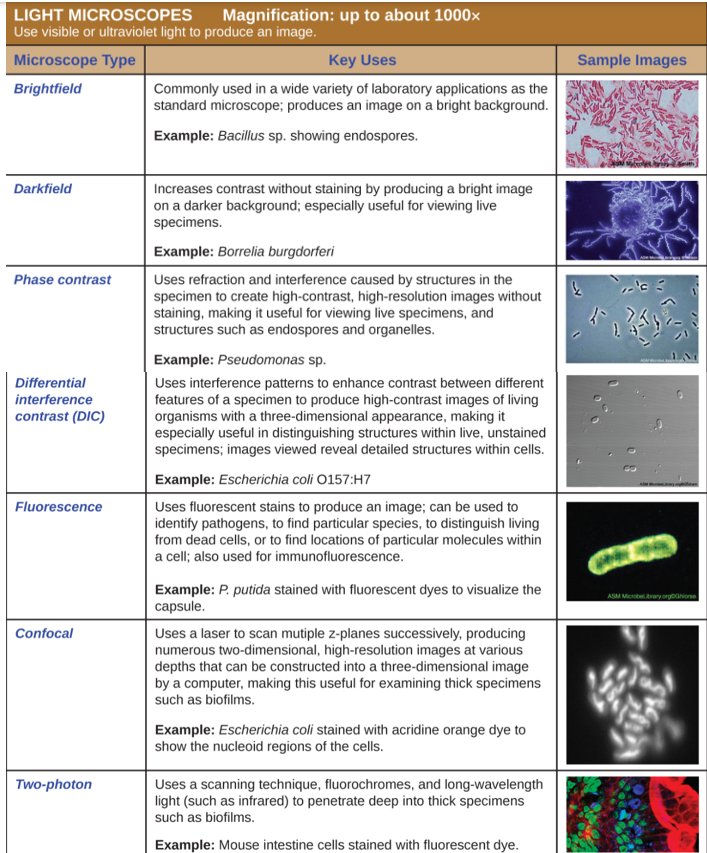
Light Microscopy
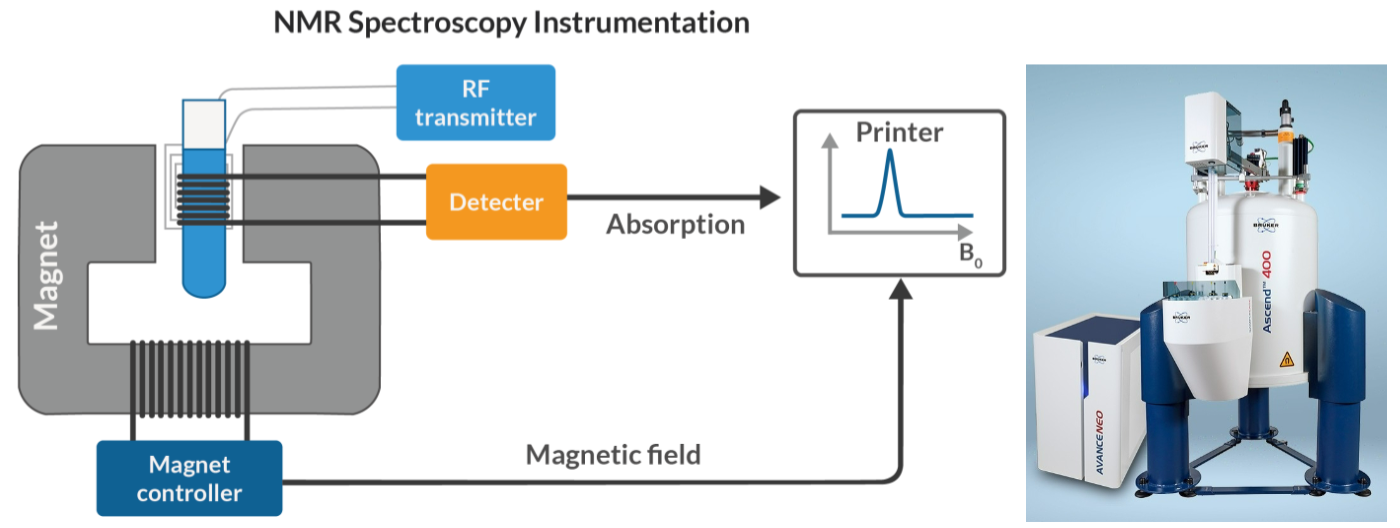
Nuclear magnetic resonance (NMR)
nuclei in a strong constant magnetic field disturbed by weak oscillating magnetic field and produce electromagnetic signal with frequency of magnetic field at nucleus
MRI (Magnetic Resonance Imaging) in humans and animals
used to determine 3D structure of biomolecules such as proteins, nucleic acids, and their complexes
provides information about structure, dynamics, and interactions of these molecules at atomic level
can be performed on samples in solution, which is closer to physiological conditions of biomolecules.
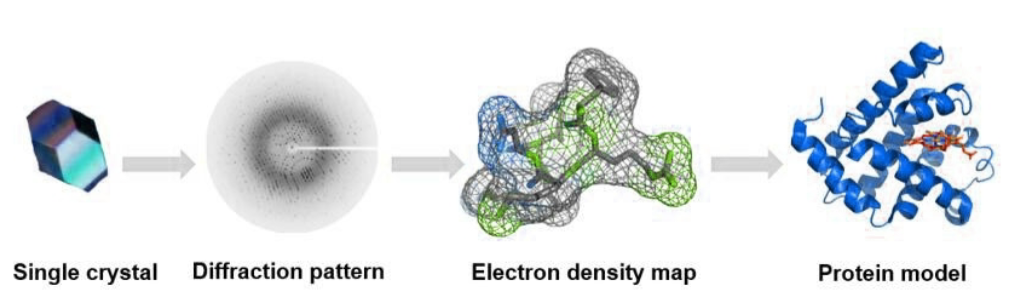
X-ray crystallography
determine 3D structure of biomolecules by analyzing diffraction pattern of X-rays passing through a crystal of molecule
provides atomic-level detail, crucial for understanding biological processes and designing drugs
diffraction pattern is analyzed to determine e- density distribution within crystal
e- density map reveals positions of atoms and allows for construction of 3D model of molecule
high resolution, ability to study small/large structures, ability to reveal fine atomic details of well-ordered macromolecules
requirement for high-quality crystals, static view of molecules, not in solution structure (crystal contacts)