Gene Control CH 16
1/39
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
40 Terms
Gene Expression Prokaryotic v. Eukaryotic
Prok: organisms regulate genes in response to environment
Euk: cells regulates genes to maintain homeostasis in organism
Regulatory Proteins
bind to DNA, regulate binding of RNA polymerase to promoter
Regulatory proteins function/structure
Bind to specific DNA sequences to control reg.
Gain access to bases of DNA at major groove + DNA motifs
Either block transcription by prevent RNA pol from binding or stimulate it by facilitating RNA pol binding to promoter
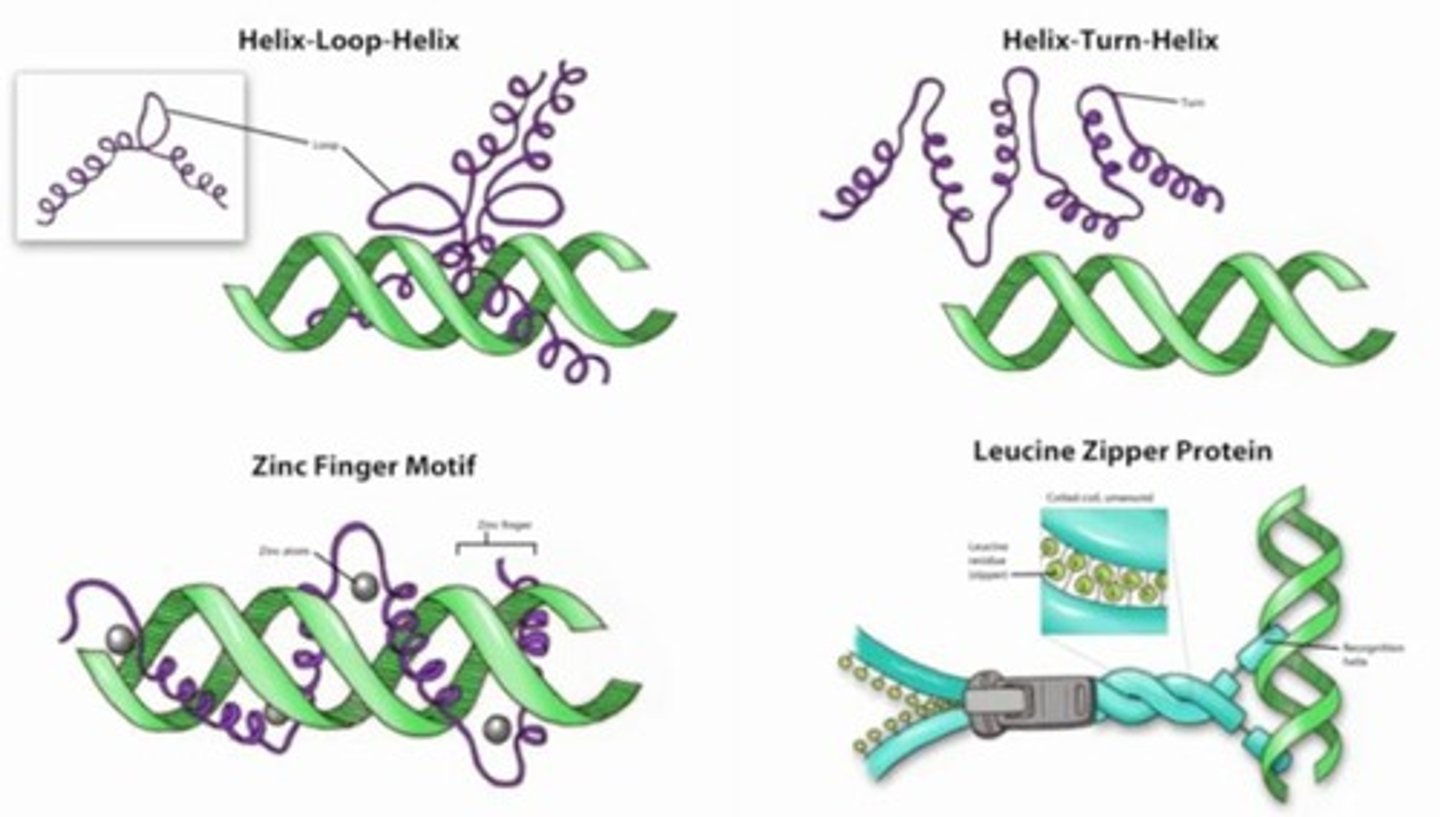
DNA-binding Motifs
Helix-turn helix motif: two a-helical segments linked by nonhelical segment
Homeodomain is special call and is critical in euk. Development
Zinc finger motif: several forms, use zinc atoms to coordinate DNA binding
Leucine Zipper motif: dimerization motif which region in on subunit interacts with similar region on another subunit forming a zipper-like connection
Prokaryotic Regulation
Transcription initiation is pos or neg controlled
positive: increases frequency (activators enhance binding of RNA pol to promoter)
Negative: decreases frequency (repressors bind to operators)
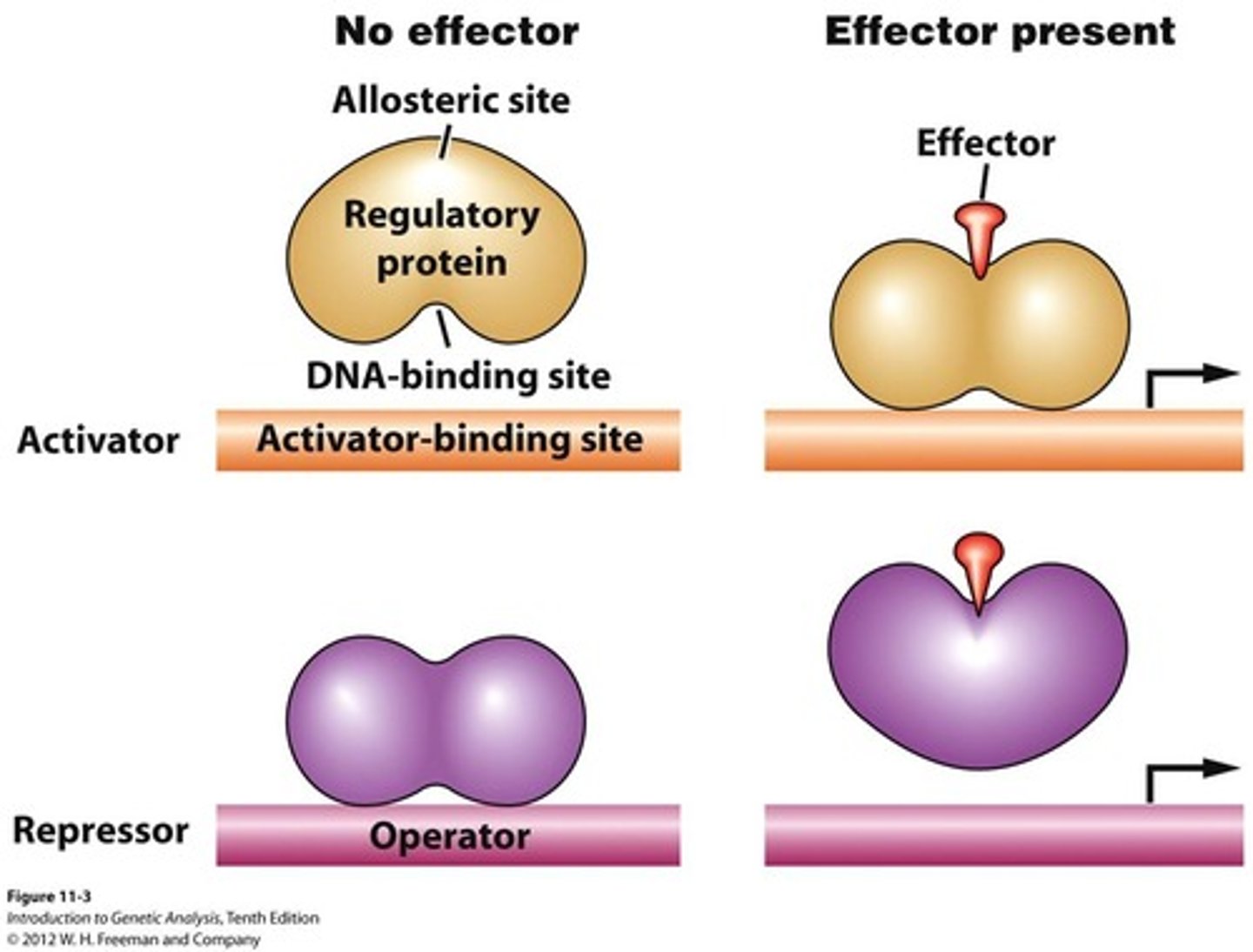
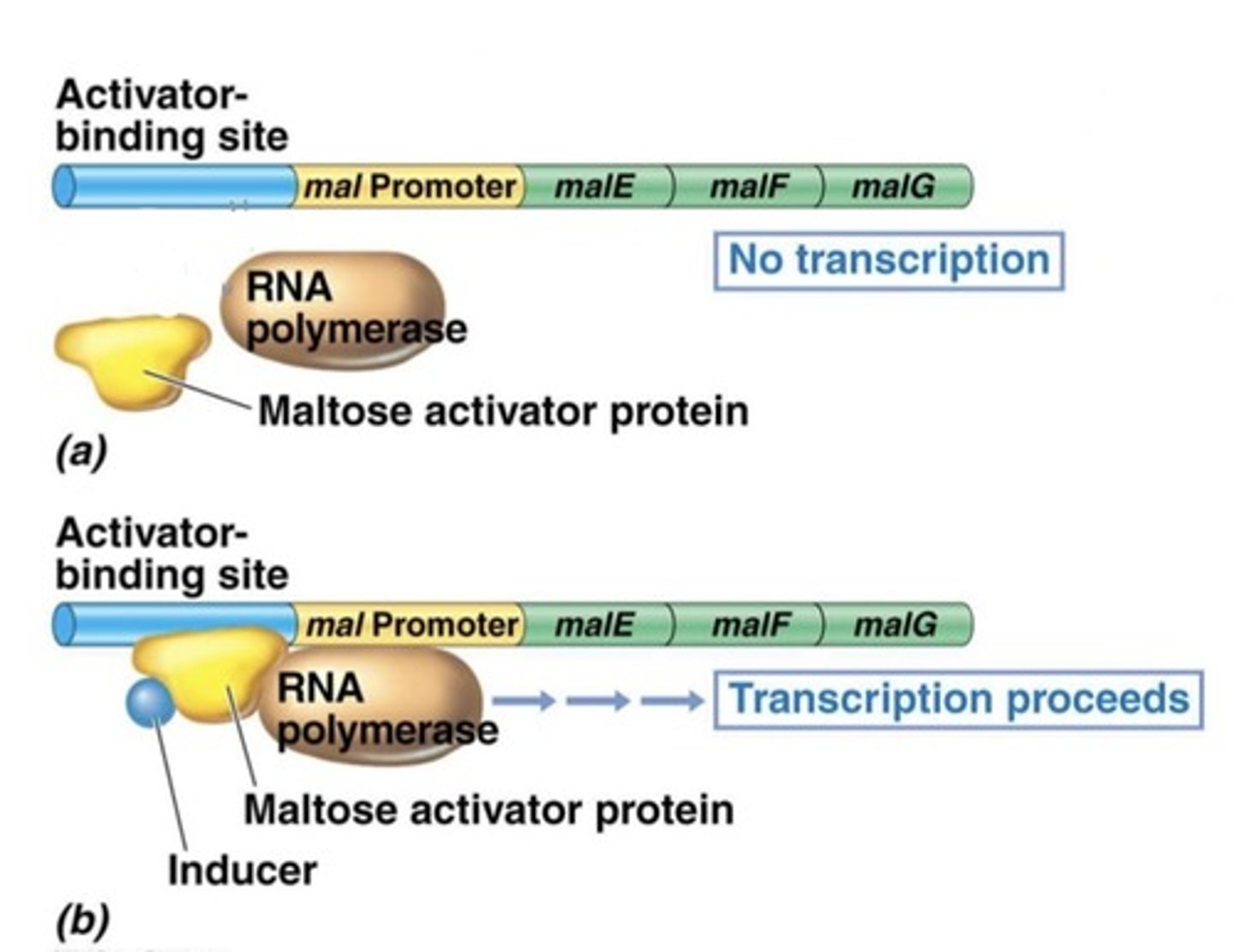
Positive control regulation
Activators enhance binding of RNA pol to promoter
increases frequency of transcription
Negative Control regulation
Repressors bind to operators (reg. cites on DNA) that prevent/decrease initiation frequency (by preventing RNA Pol)
decrease frequency of transcription
Effector Molecules (regulation)
Substances that influence activators and repressors. (inducers)
Induction (prok)
enzymes for certain pathways are produced in response to a substrate
Inducers: bind to repressor proteins→ allows RNA pol to bind now
Repression (prok)
Gene expression decreases despite potential enzyme production.
Repressor proteins: bind to operator preventing transcription (sometimes need effector molecule to remain bonded)
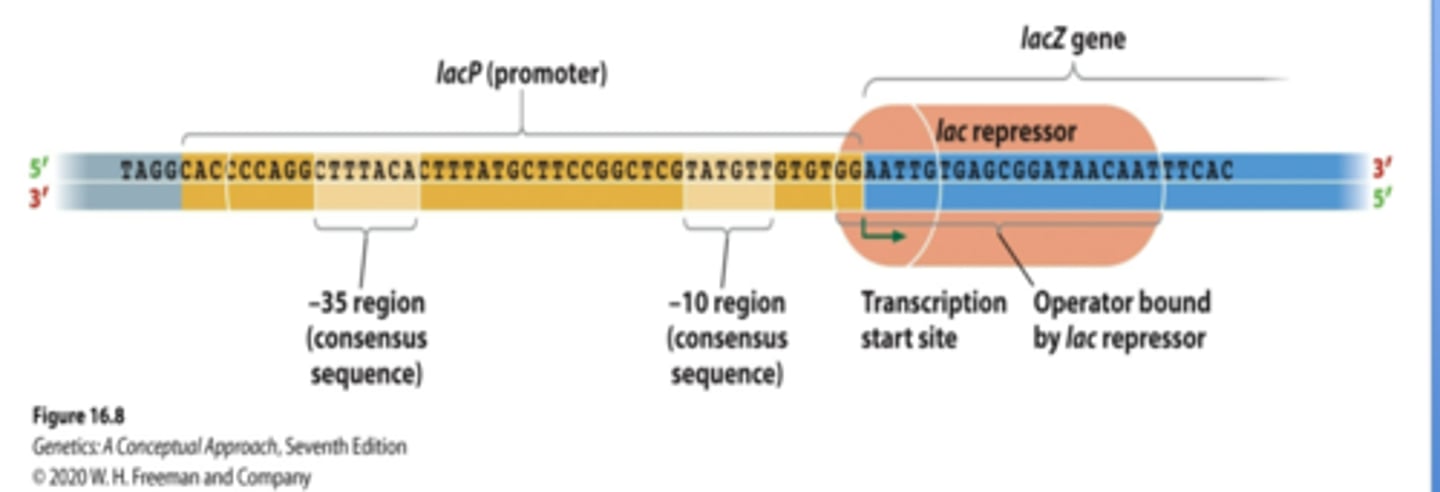
Lac Operon
Encodes proteins necessary for use of lactose as energy storage (when glucose is short)
lac Z,Y,A
lac repressor lac L
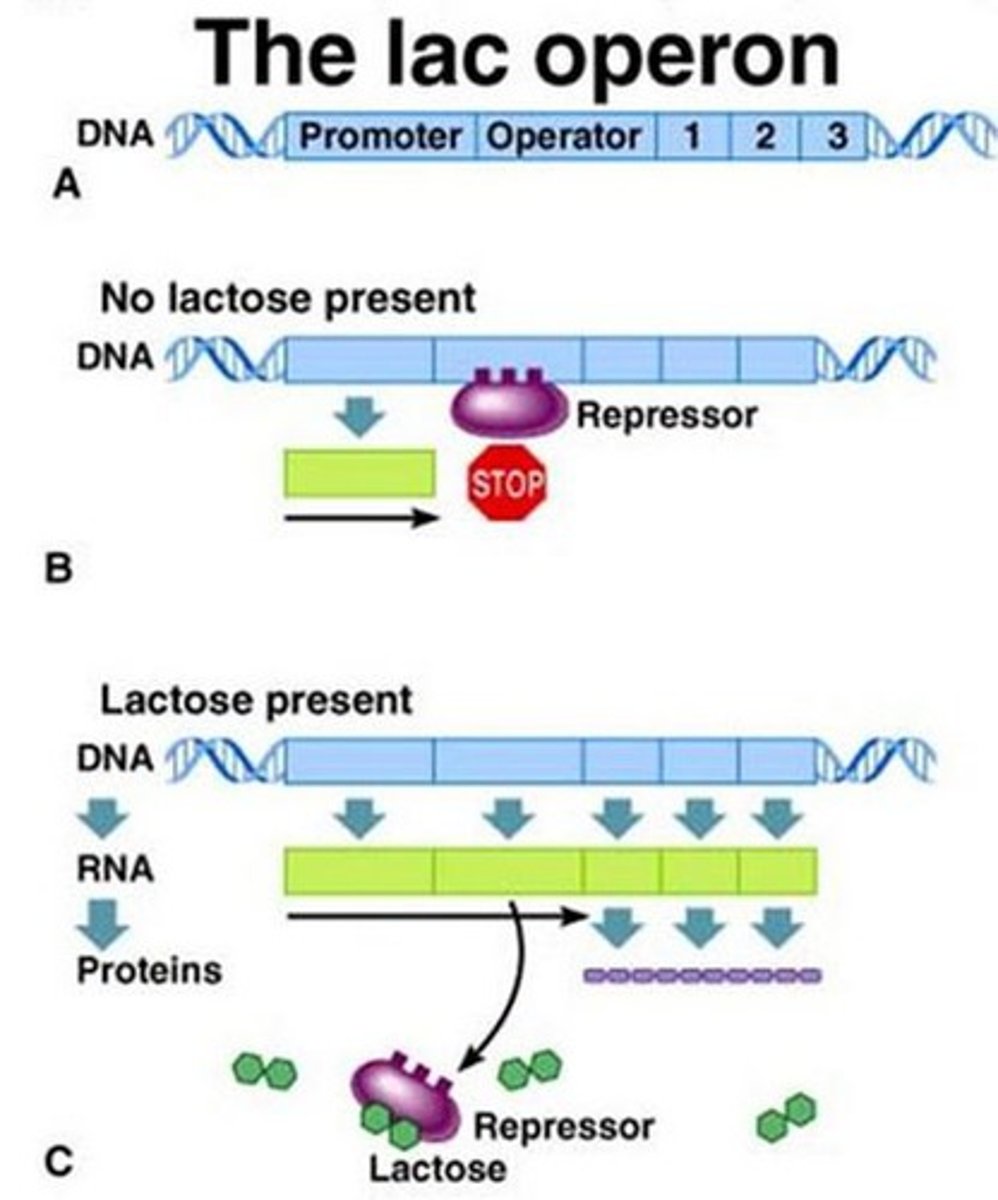
Lac Repressor
Protein that inhibits transcription of the lac operon.
Allolactose
Inducer that prevents lac repressor binding.
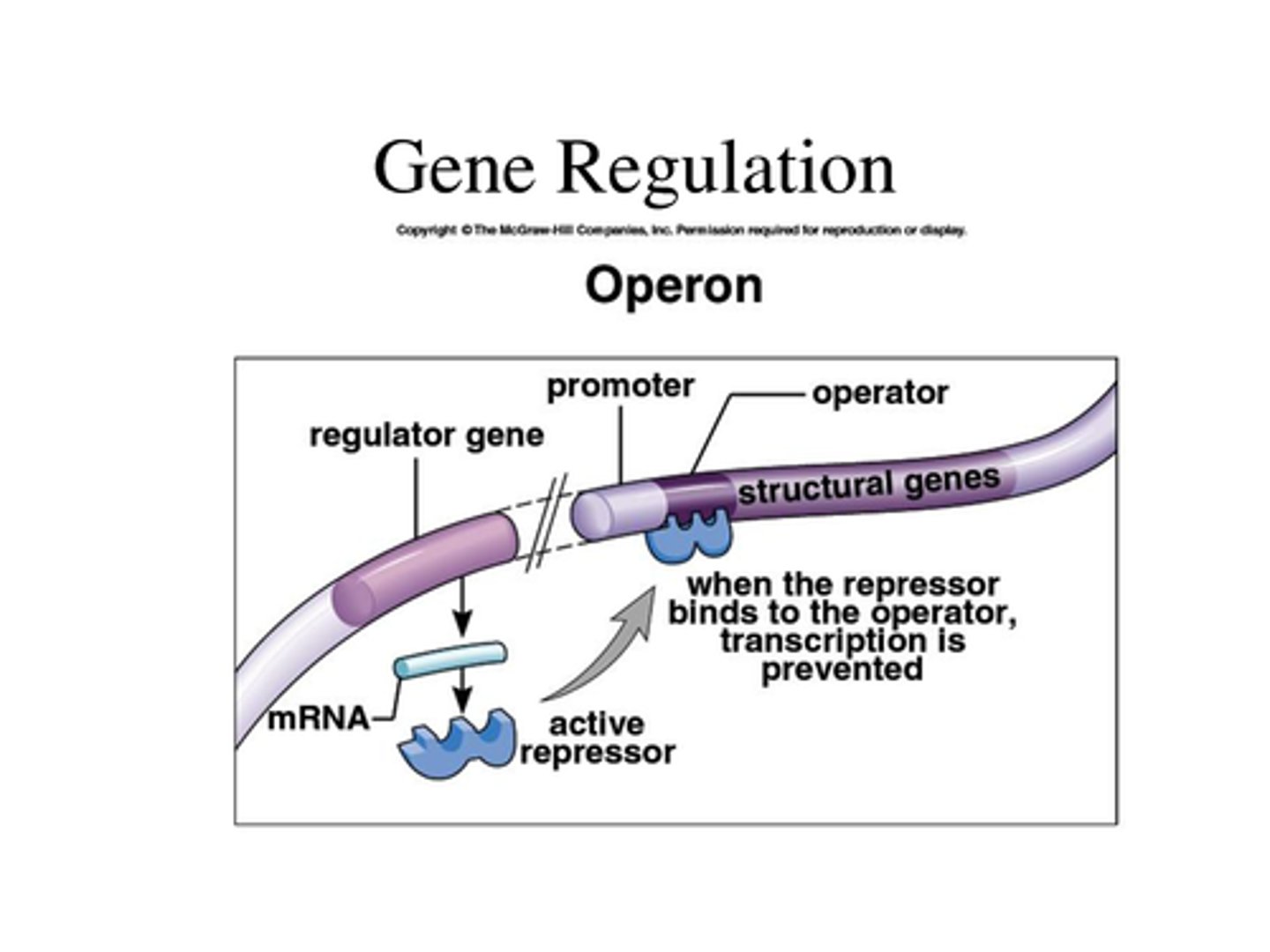
Negative Regulation of lac operon
Lac repressor binds to operator to block transcription
In the presence of lactose, an inducer molecule (allolactose) binds to repressor protein
Repressor can no longer bind to operator
Transcription proceeds
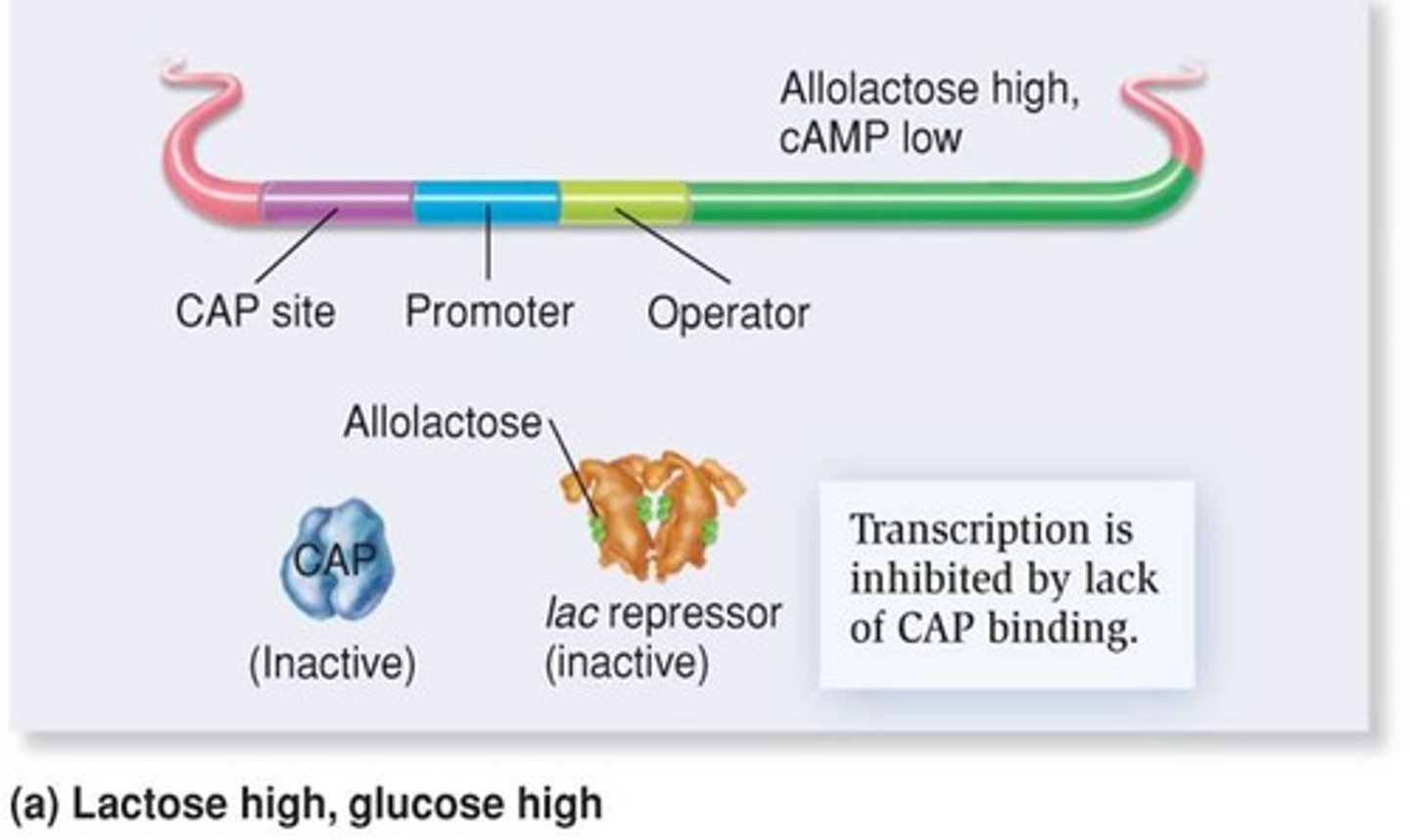
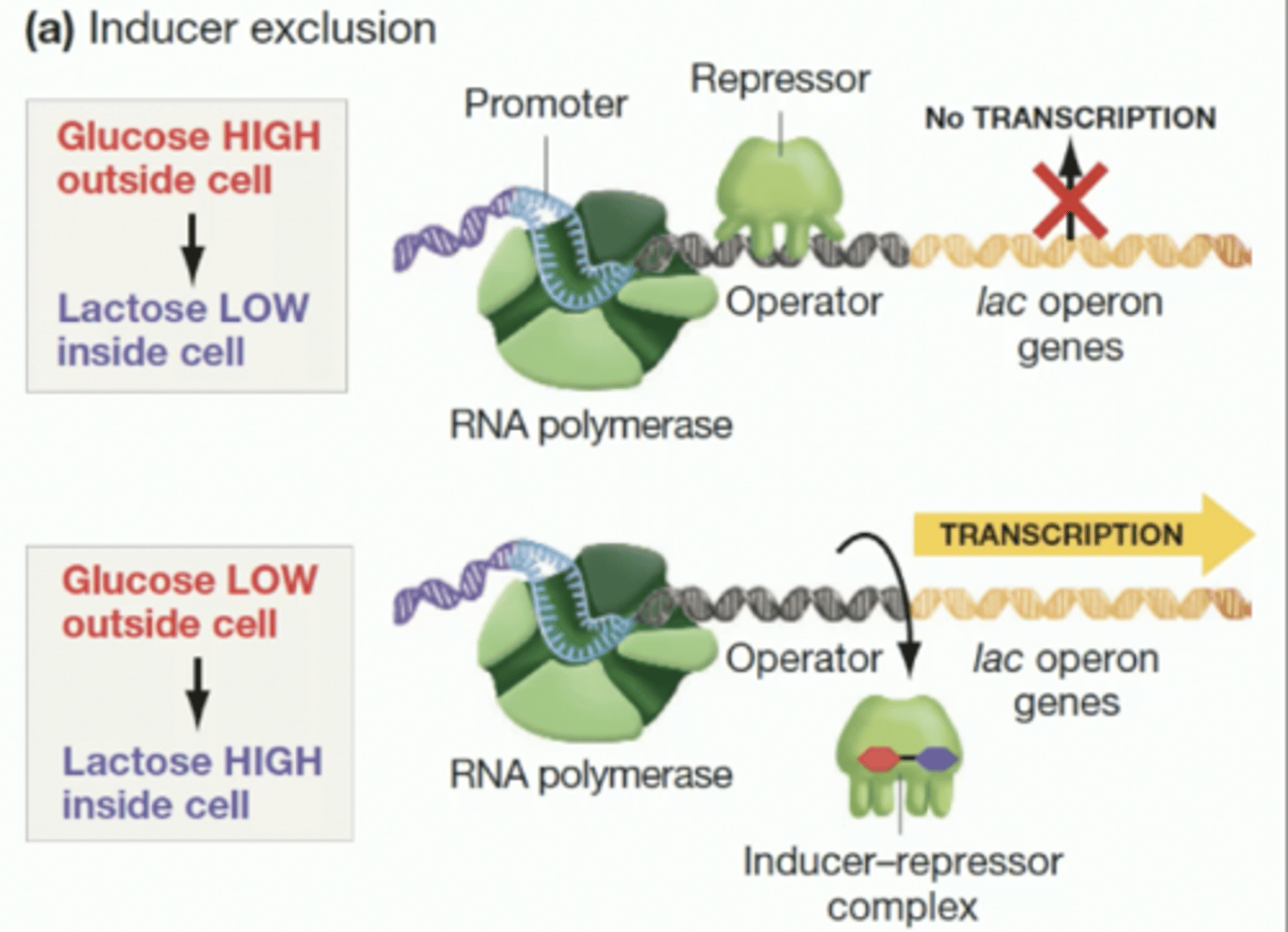
Glucose Repression
Preference for glucose over other sugars in metabolism.
Catabolic Activator Protein (CAP): an allosteric protein with cAMP as effector
Level of cAMP in cells is reduced in presence of glucoses if not stimulation or transcription from CAP-responsive operons takes place
Inducer Exclusion
Presence of glucose inhibits lactose transport into the cell.
Operons
Gene clusters regulated together in prokaryotes.
Operator
DNA region where repressors bind to block transcription.
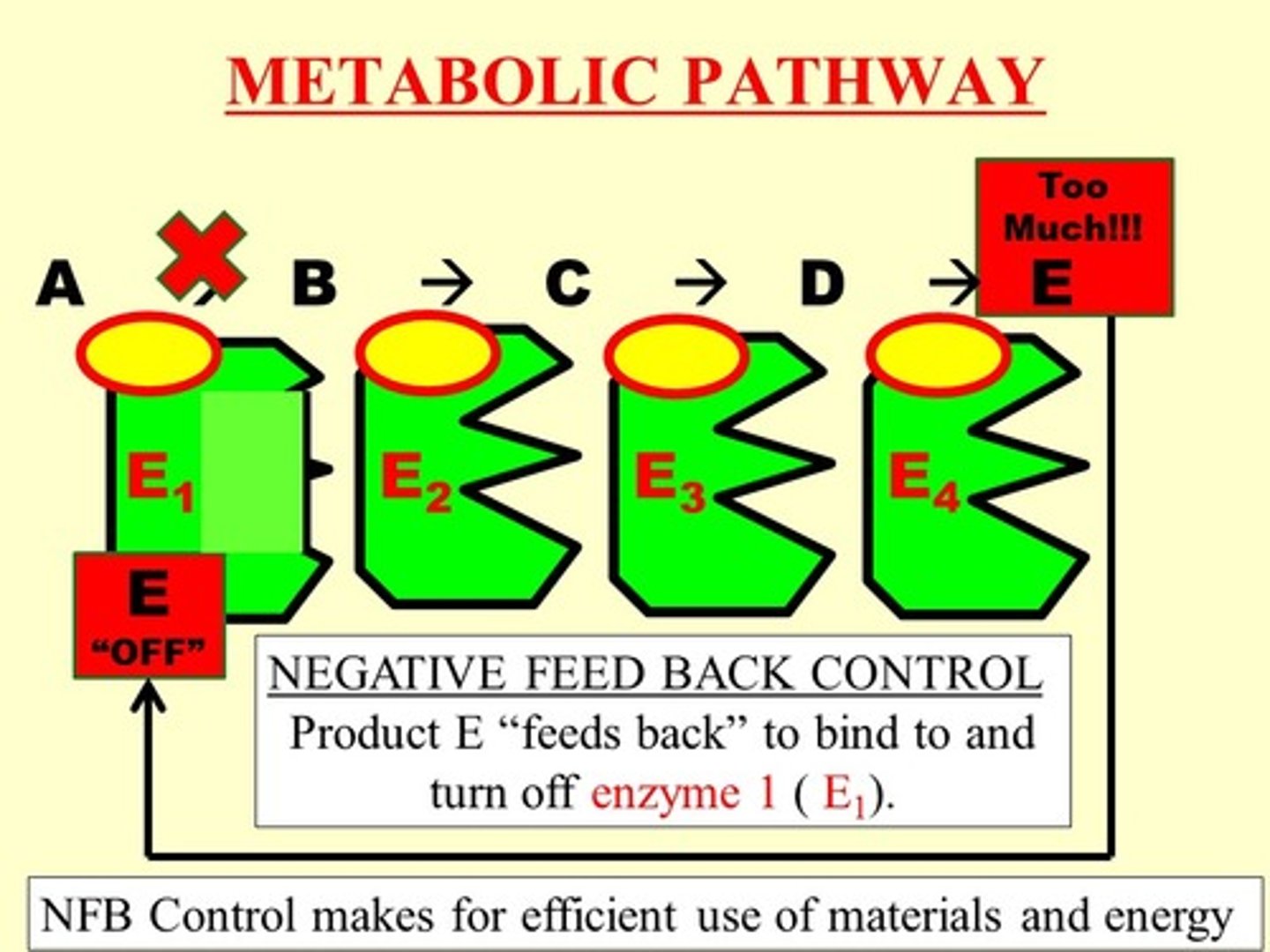
trp Operon
Operon encoding tryptophan biosynthesis genes. (important for making proteins)
Operon is not expressed when cell contains sufficient amount s of tryptophan
Operon is expressed when levels of tryptophan are low
Trp Repressor
helix-turn-helix protein that bonds to operator site located adjacent to trp promoter
Negative Regulation of trp operon
Trp repressor binds to operator to block transcription
Binding of repressor to operator requires a co-repressor which is tryptophan (operon is repressed)
When tryptophan levels fall, repressor cannot bind to operator (operon id depressed, versus being induced)
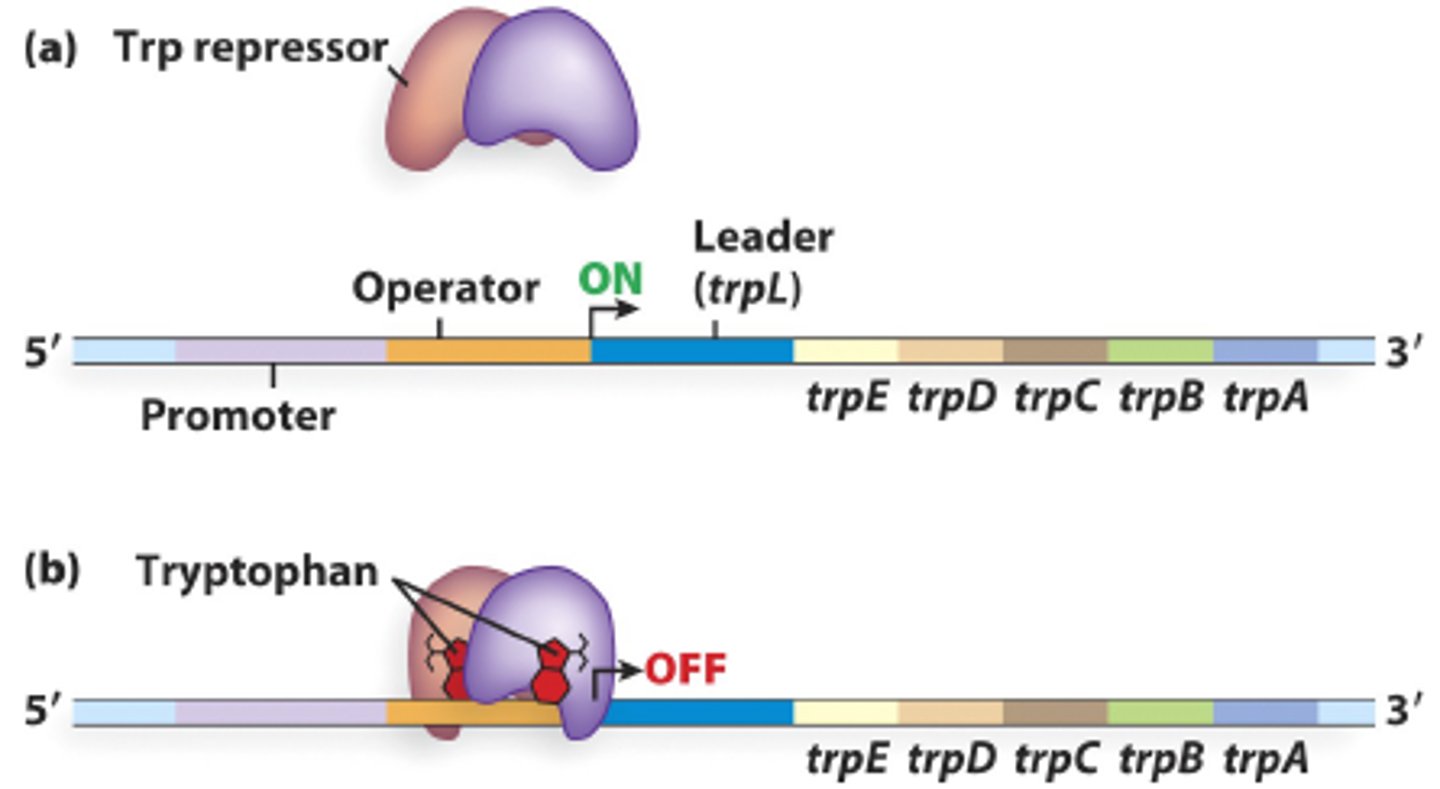
Co-repressor
Substance required for repressor binding to operator. (trp operon)
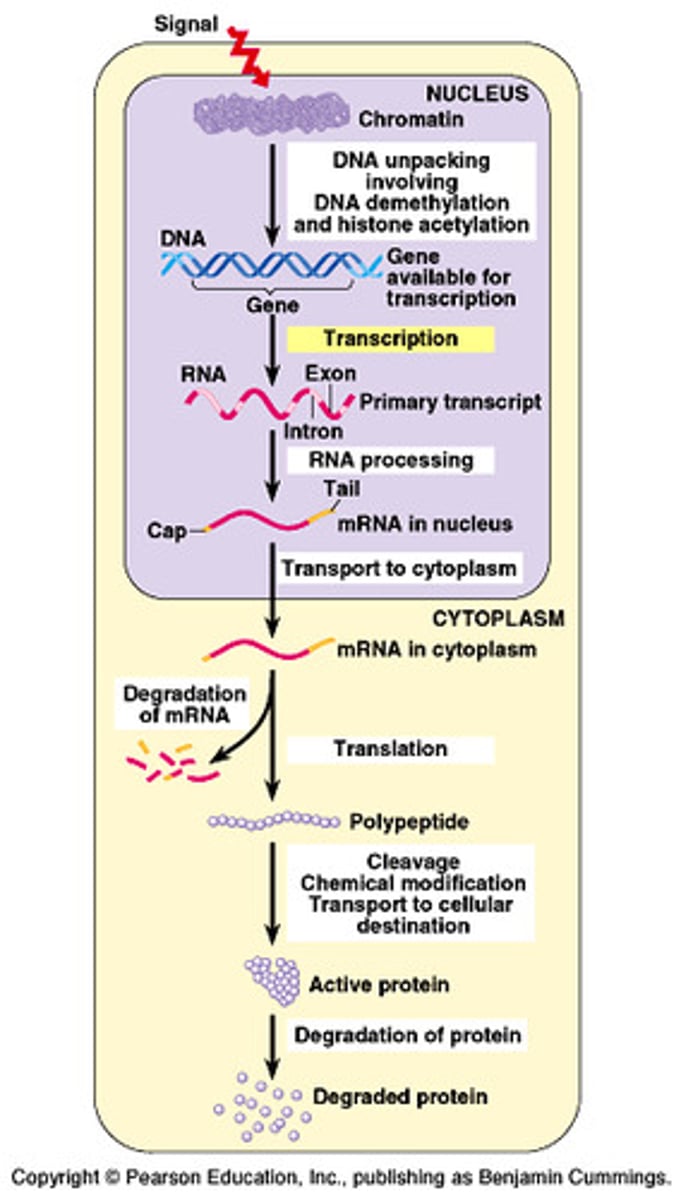
Eukaryotic Regulation
More complex; major differences from prokaryotes...
Euks. have DNA organized into chromatin (complicates protein-DNA interaction)
Euks. transcription occurs in nucleus while translation occurs in cytoplasm
Amount of DNA involved in regulating euks. genes is much larger
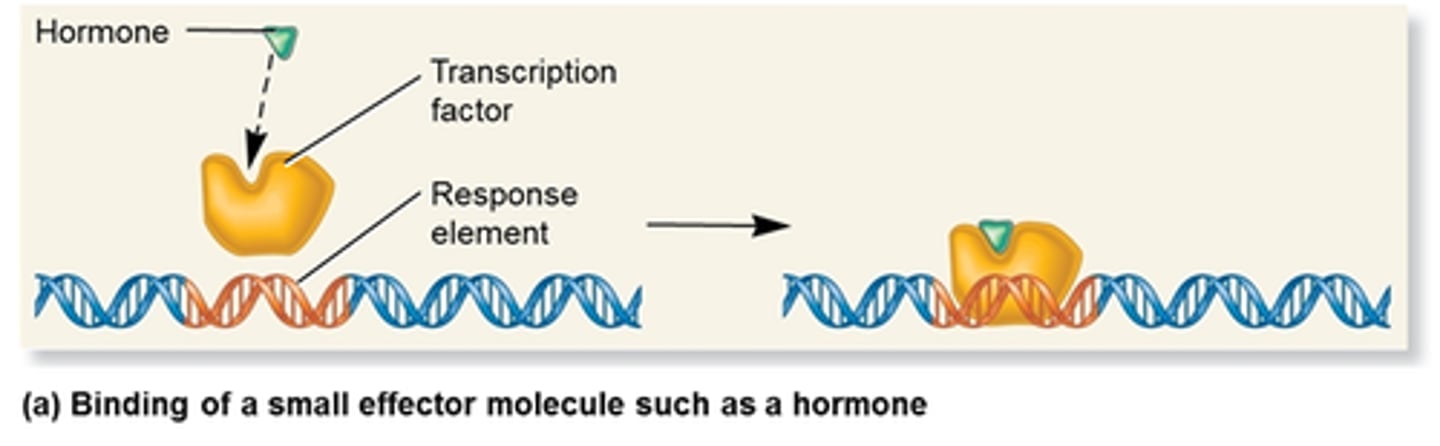
Transcription Factor Nomenclature
General transcription factors are named with letter designating following TF, & roman numeral designation of which RNA pol the factor interacts with
Ex. TFI-> transcription factor RNA pol I
Ex. TFIID-> transcription factor pol II D
General Transcription Factors
Proteins necessary for transcription, but only do so at the basal level.
TFIID recognizes TATA box sequences
After TFIID binds TFIIE, TFIIF, TFIIA, TFIIB, and TFIIH bind + many transcription-association factors = TAFs
This initiation complex can initiate synthesis at a basal level
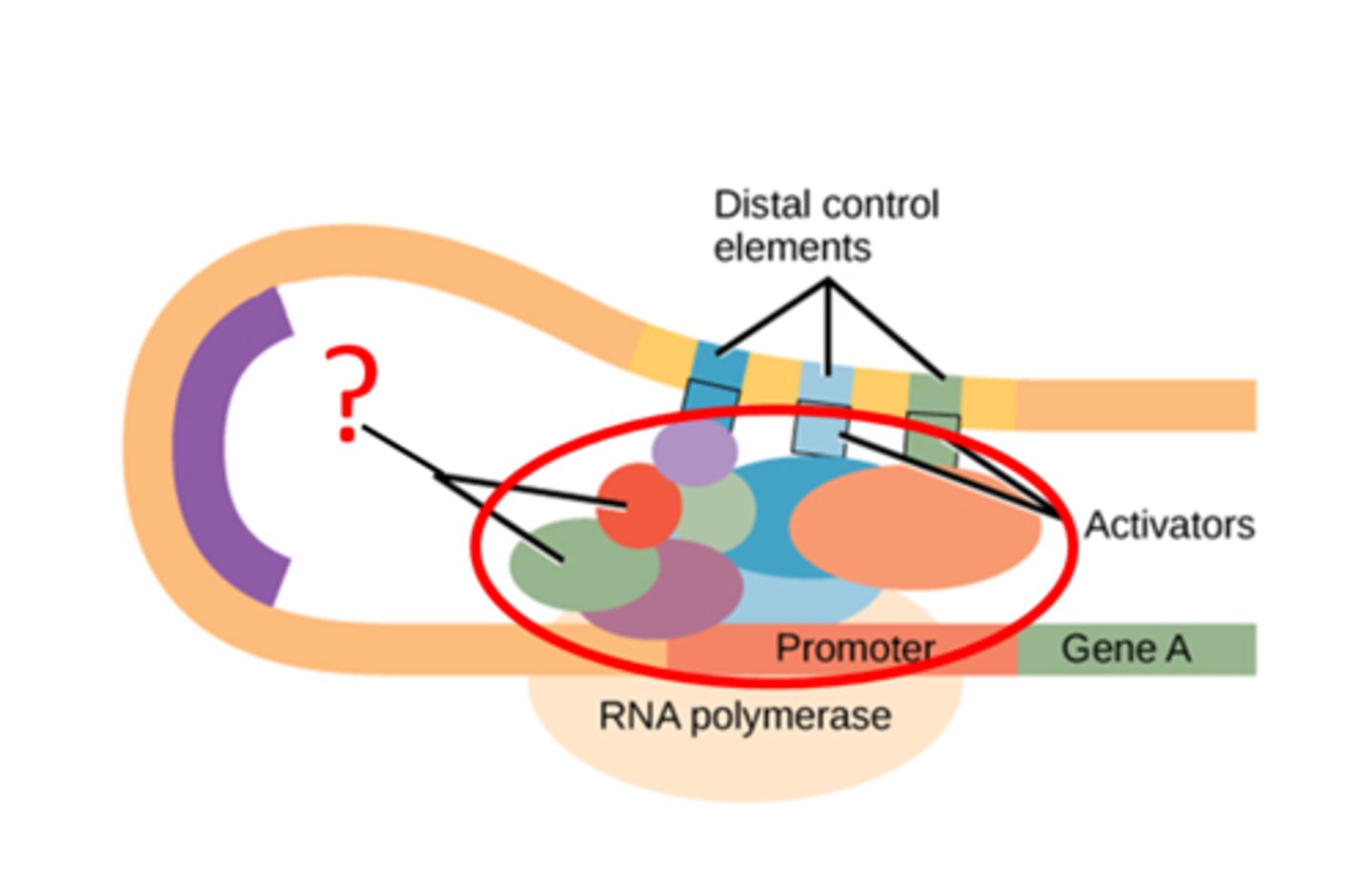
Specific Transcription Factors
Factors that enhance transcription in specific conditions, stimulation higher levels
Each factor consists of DNA-binding domain & separate activating domain that interacts with the transcription apparatus
These domains are independent in the protein
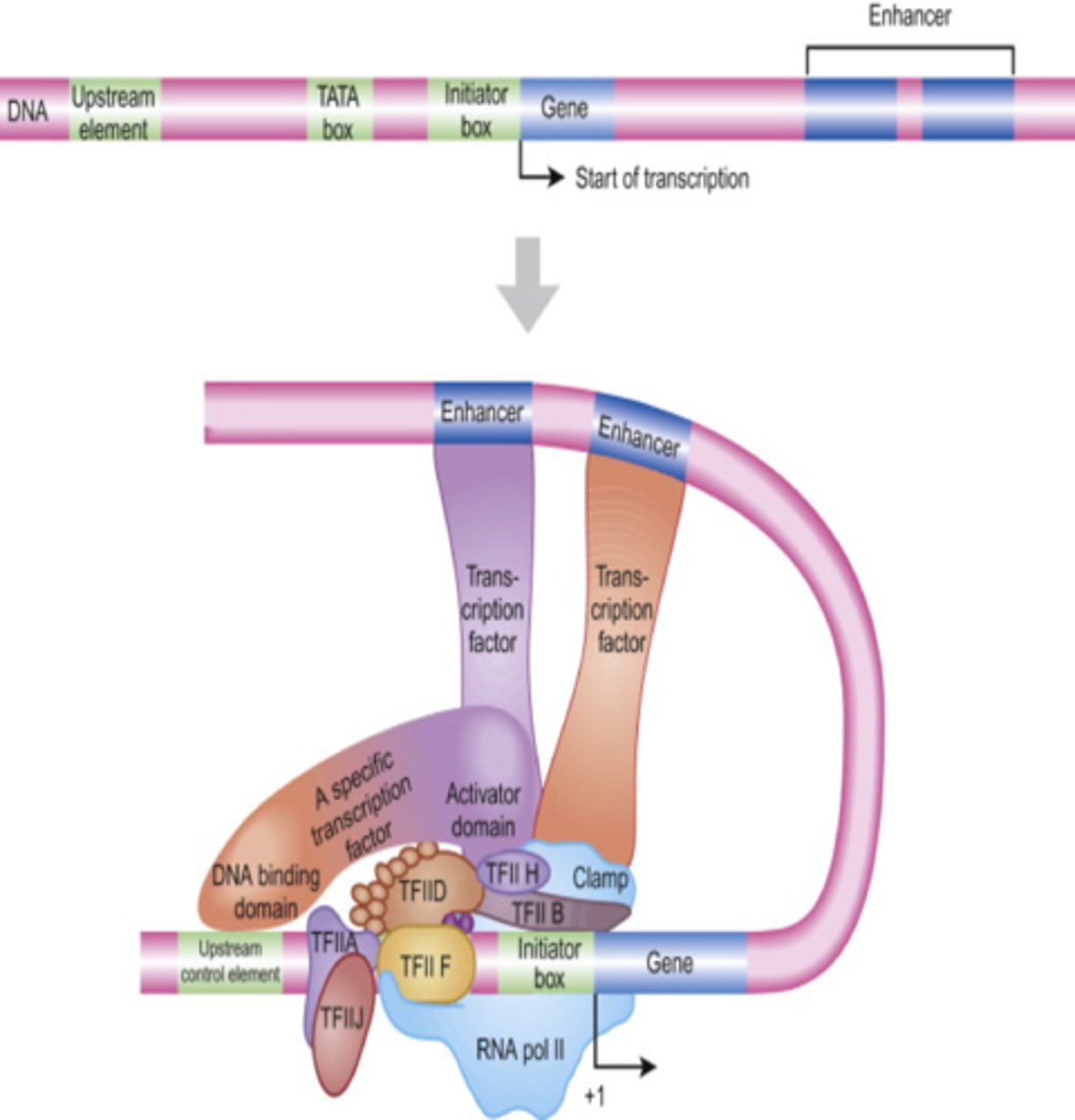
Promoters
DNA sequences where transcription factors bind.
Mediate binding of RNA polymerase II to promoter
Enhancers
binding site of specific transcription factors
Act over large distances by bending DNA to form loop to position enhancer closer to promoter
Activator protein binds to it + mediator= initiates transcription
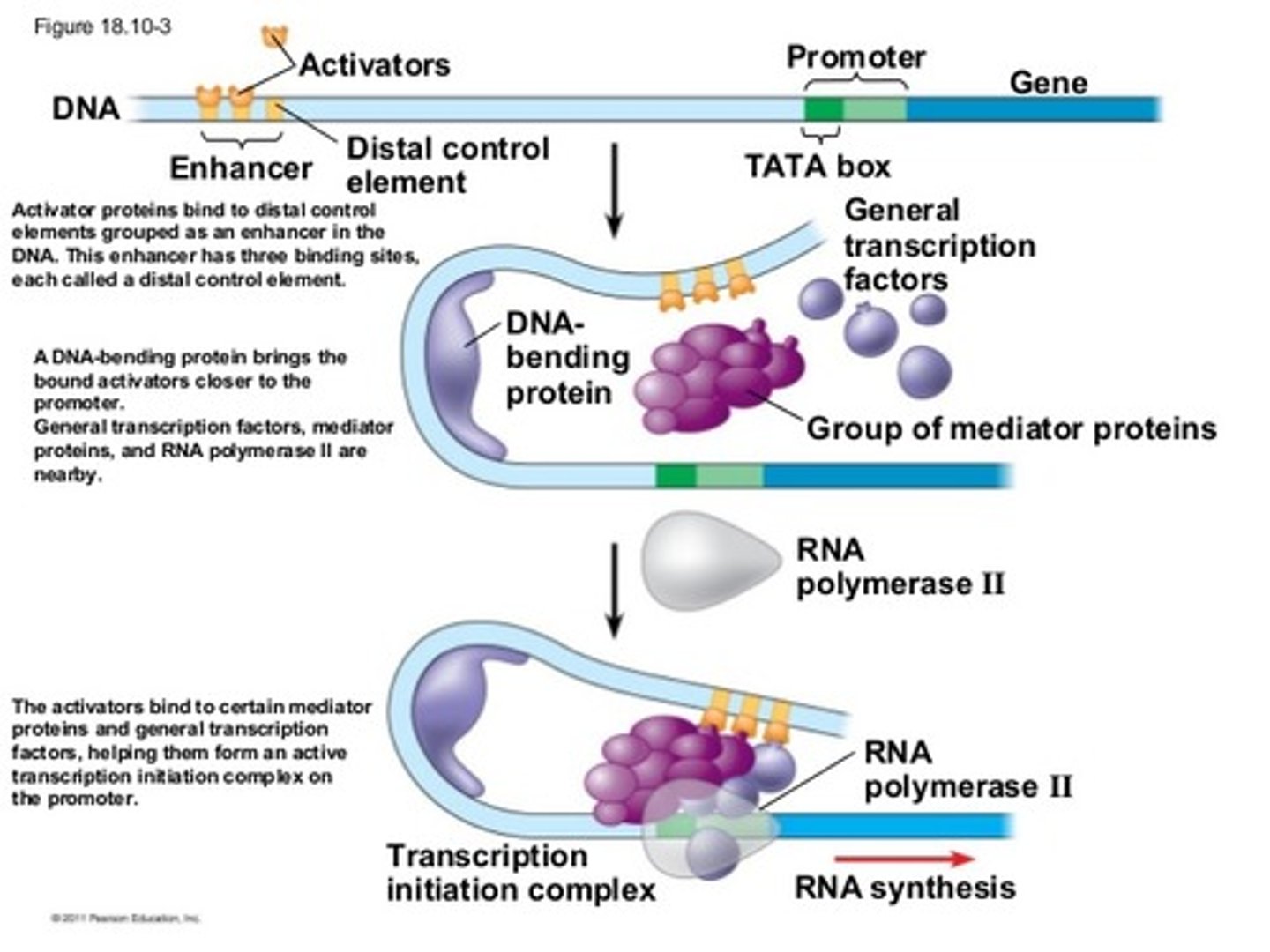
Coactivators & mediators
Coactivators and mediators are also required for the function of transcription factors
Bind to transcription factors and bind to other parts of the transcription apparatus
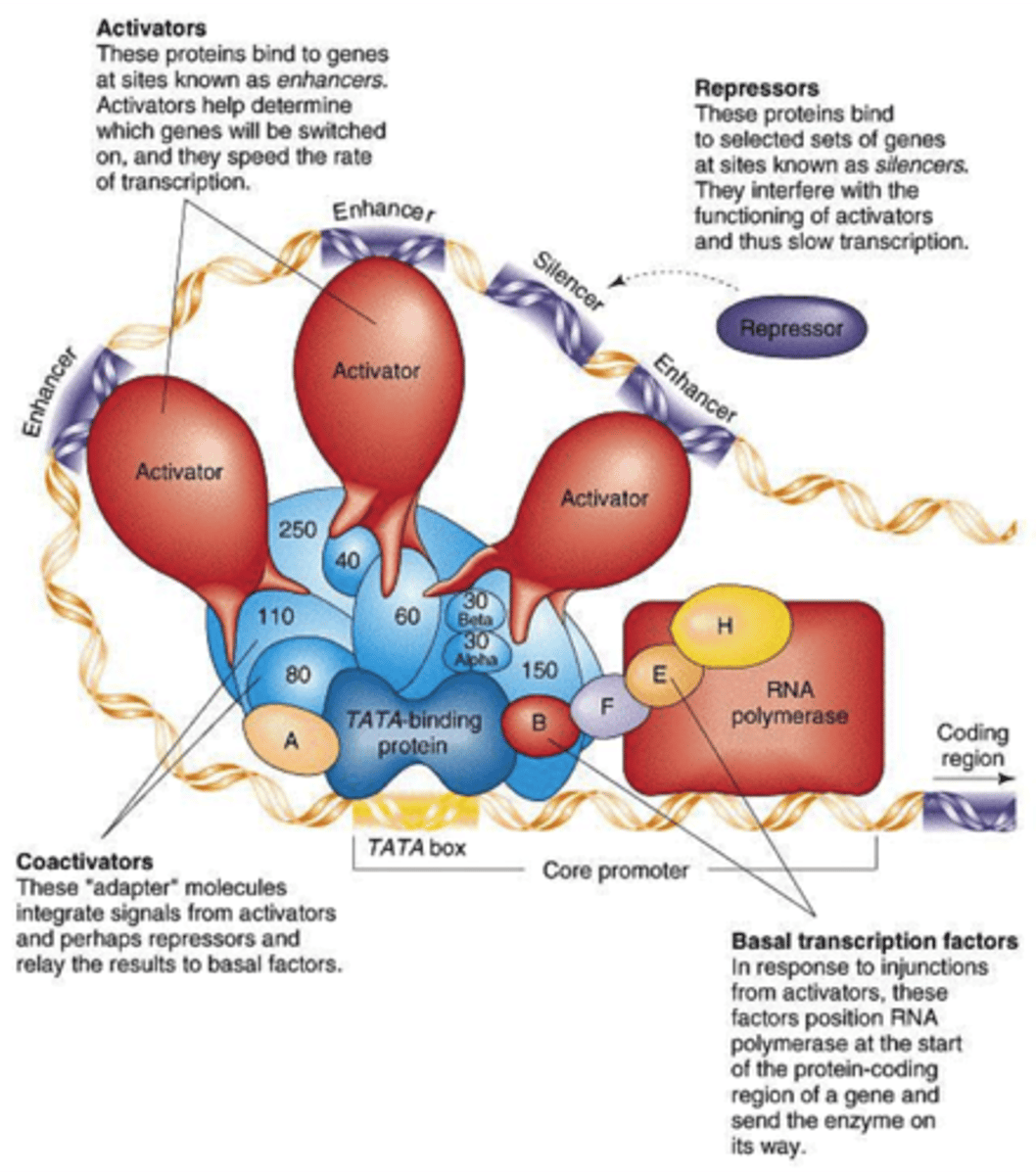
Transcription complex
Virtually all genes transcribed by RNA pol II need the same suite of general factors to assemble initiation complex
General and specific TFs near promoter were Pol II binds + activators connecting enhancers to transcription apparatus
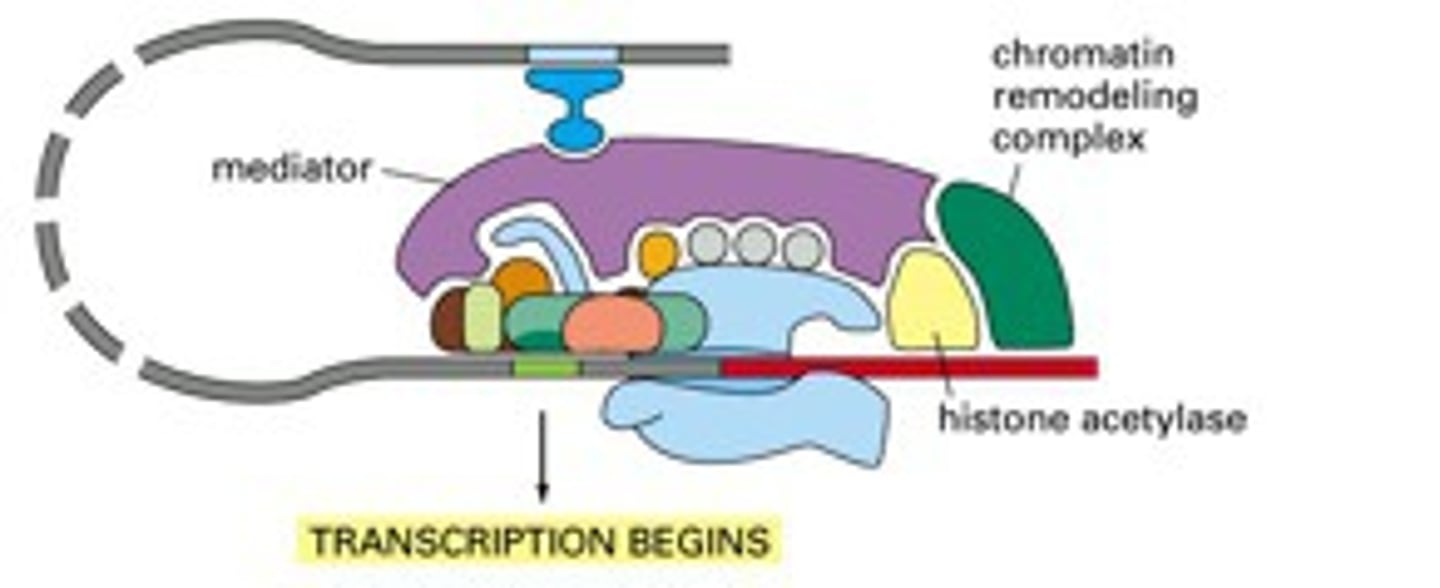
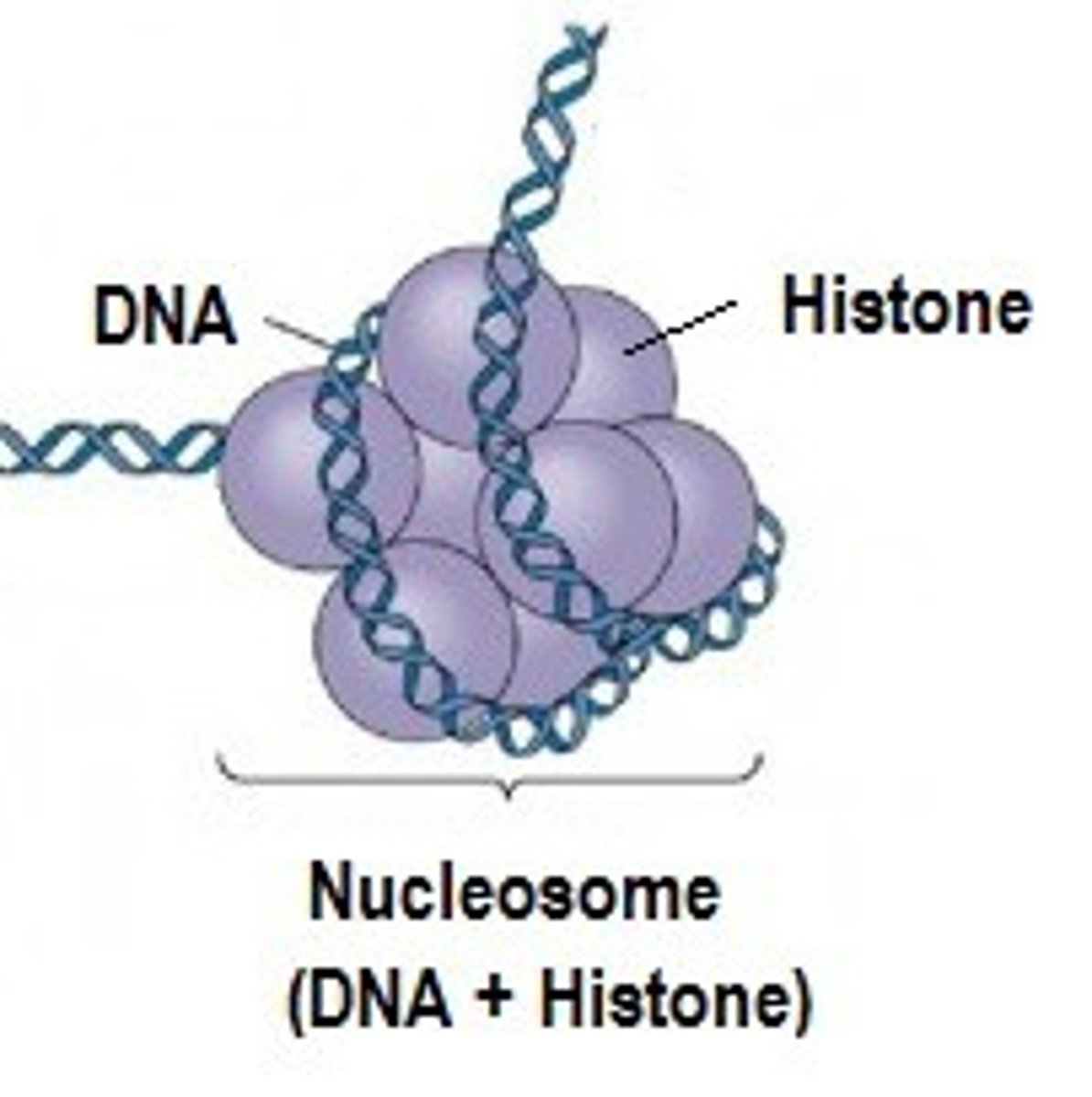
Chromatin Structure (euk)
DNA are wound around histone proteins to form nucleosomes
Nucleosomes & histones complicates process of transcription (restricts access of transcription machinery to the DNA)
Chromatin structure is selectively modulated to allow transcription, etc.
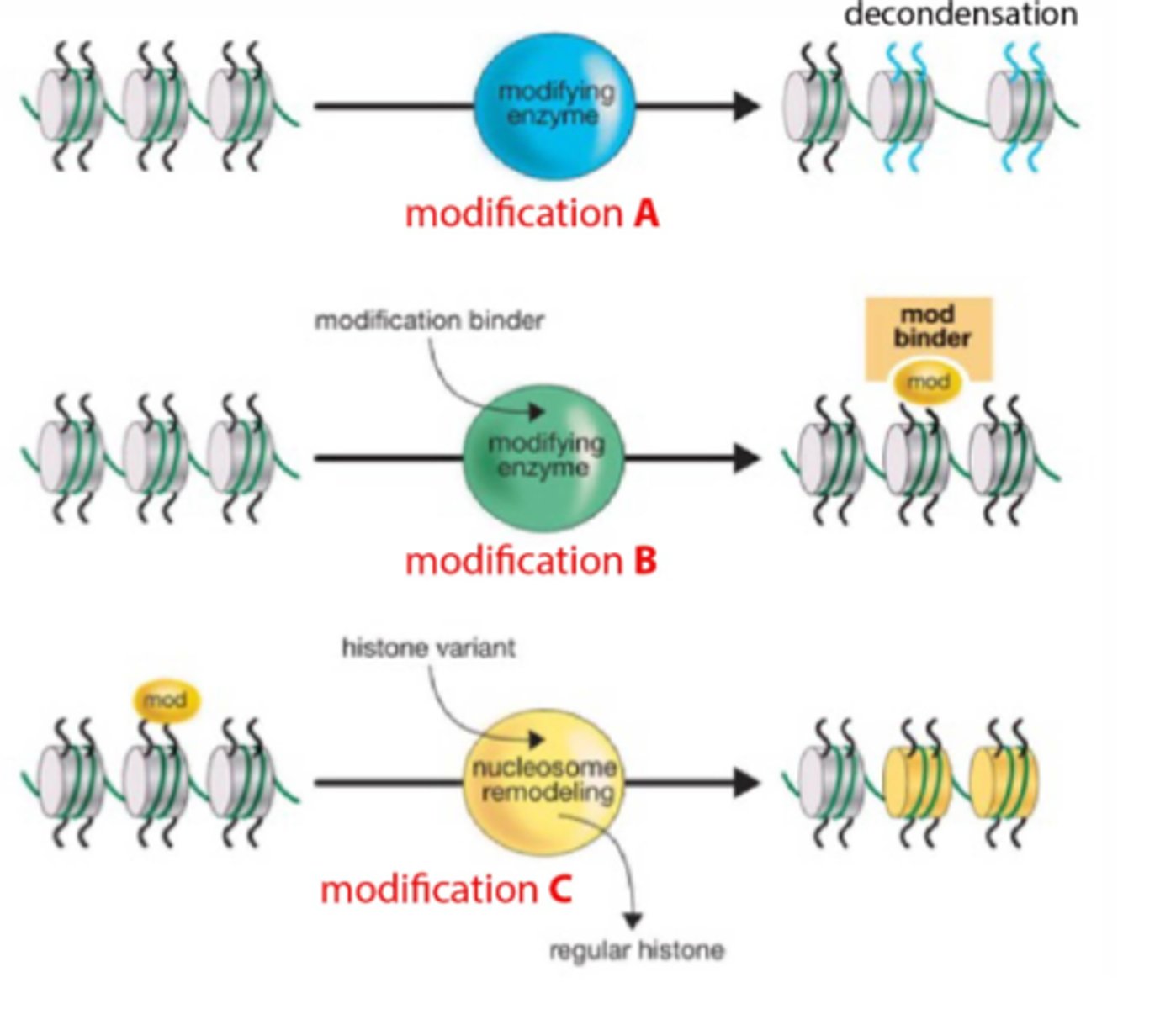
Chromatin modifications
DNA Methylation
High levels of DNA methylation correlate with inactive genes
Allele-specific gene expression seen in genomic imprinting is least partially due to DNA methylation
X-chromosome inactivation
Mammalian females inactive one X chrom. As a form of dosage compensation
(X-inactivation-specific transcript (Xist) coats entire inactive X chrom. Leading to histone modification )
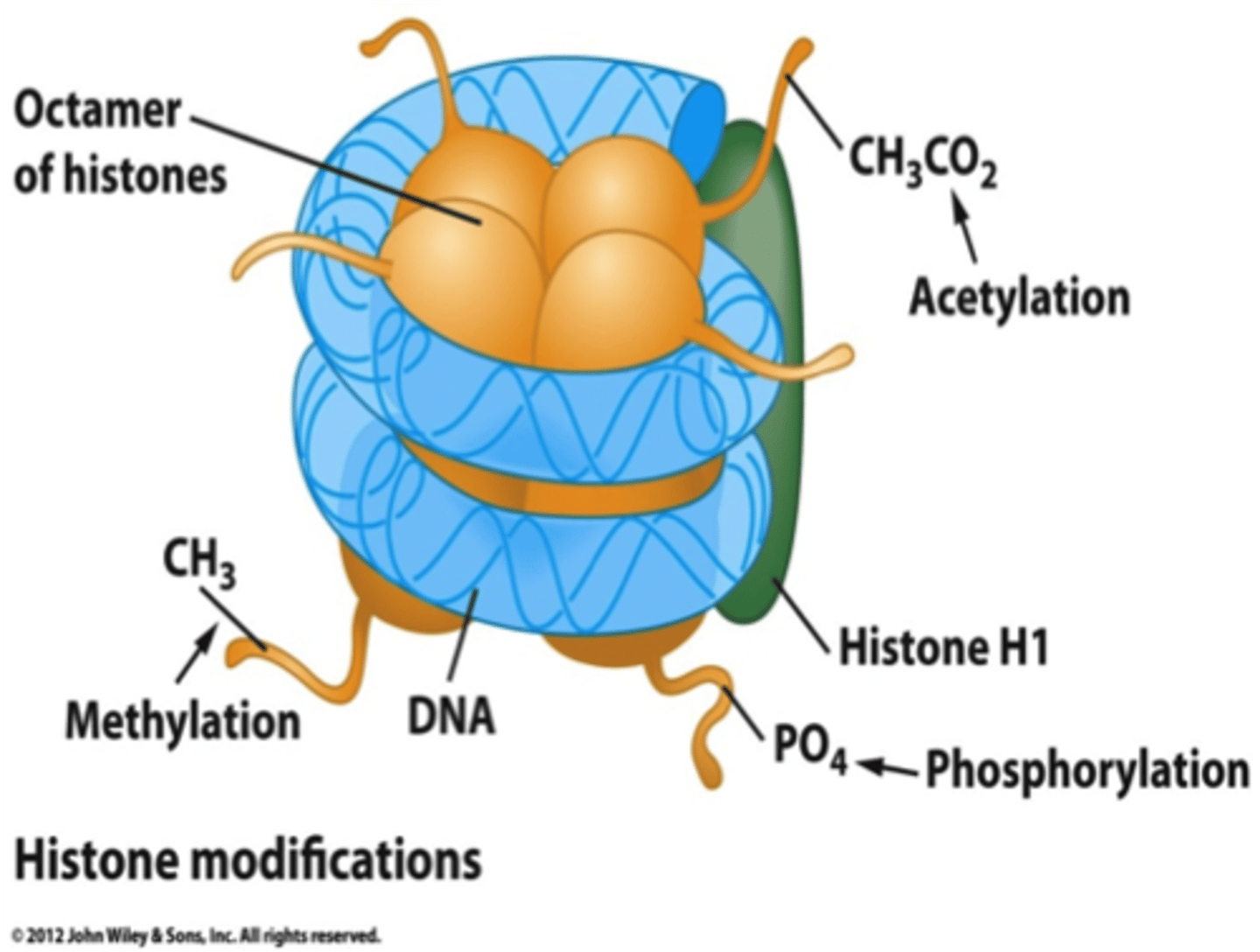
Histone Modification
Four possible histones can be modified
Acetylation, methylation, phosphorylation are all possible modifications
In general acetylation is correlative with active sites of transcription
histone acetylases (HATs) (Transcription is increased by removing higher-order chromatin structure preventing transcription )
Histone deacetylases (HDACs) (remove acetyl groups from histones)
Chromatin-Remodeling Complexes
contain enzymes that modify histones & DNA + alter chromatin structures
ATP-dependent chromatin remodeling factors (one class of remodeling factors)
Function as molecular motors
Catalyze 4 diff. Changes in DNA/Histone binding
Make DNA more accessible to regulatory proteins
Posttranscriptional Regulation (euk)
Small RNAs (miRNA and siRNA)
Alternative splicing
RNA editing
mRNA degradation
RNA-induced silencing complex (RISCP): RNA that can affect gene expression by inhibiting translation by degrading mRNAs
RNA Editing
Editing mature mRNA transcripts can produce an altered mRNA that is not truly encoded in genome
In mammals, RNA editing involves chemical modification for a base to change its base-pairing properties
(mRNA for serotonin (5HT) receptor (brain receptor for opiates) is edited at multiple sites to produce 12 diff isoforms of protein)
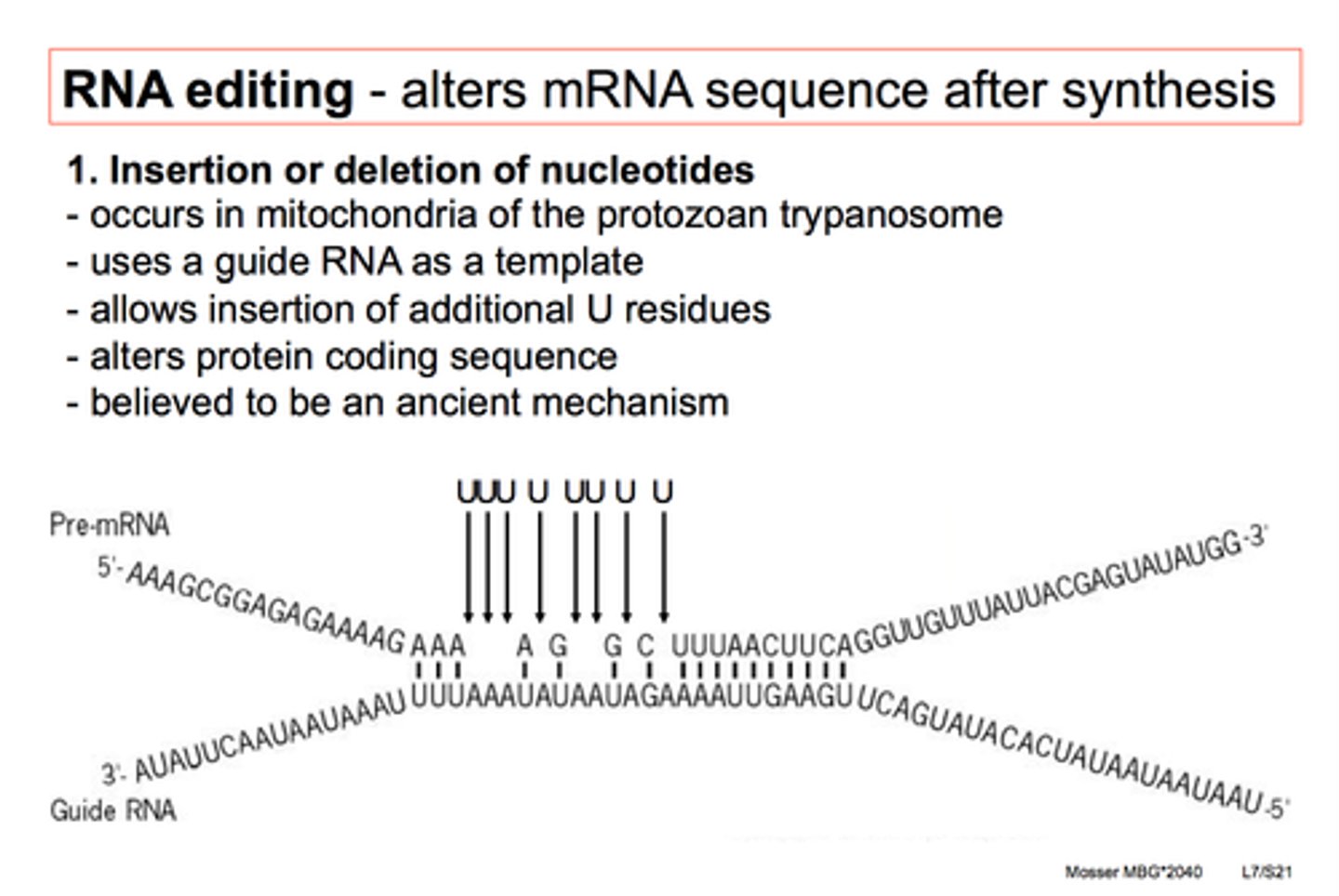
Translation Repressor Proteins
control initiation of translation by bonding to start of mRNA which prevents ribosomes from binding
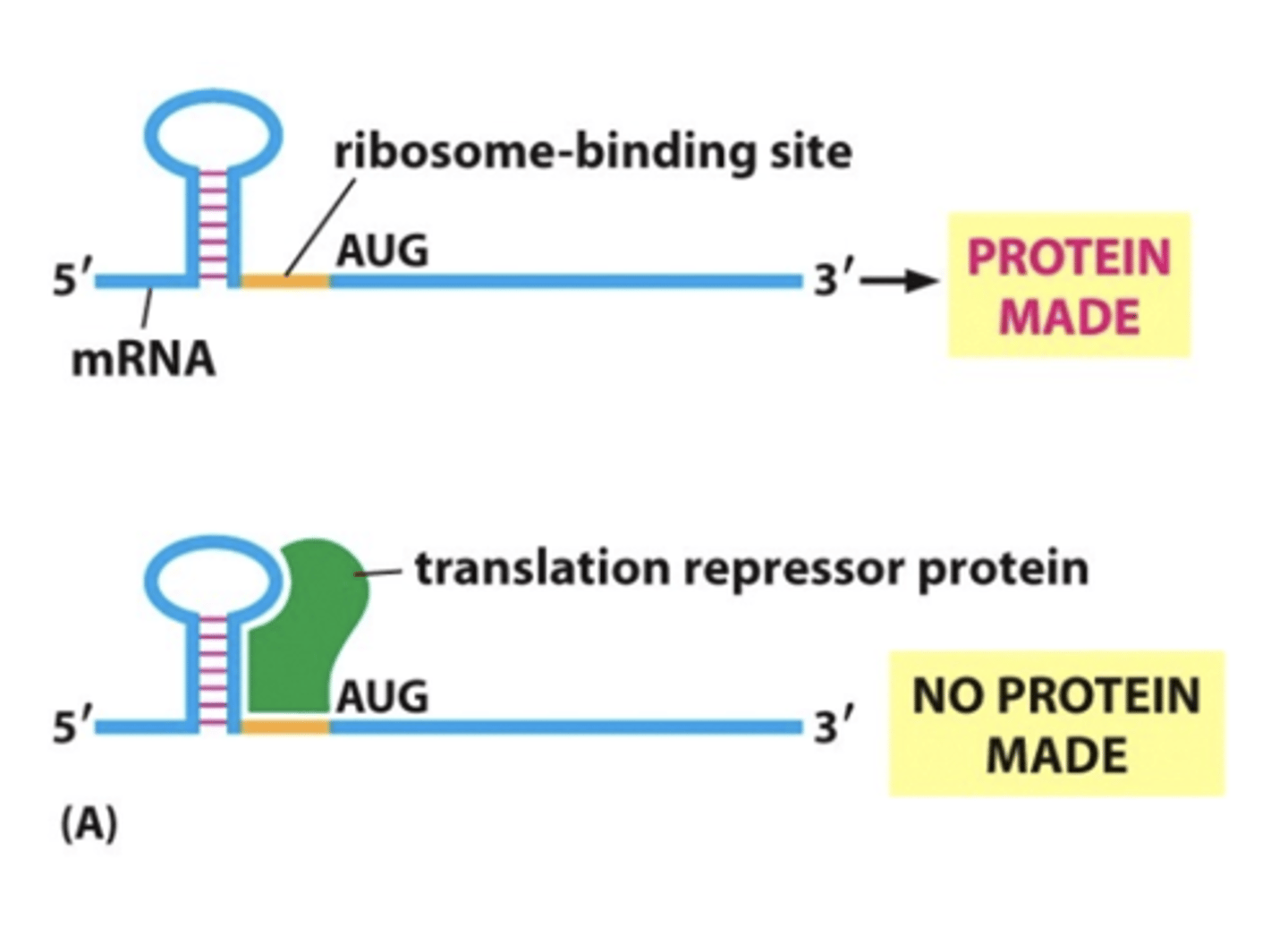
Protein degradation
Ubiquitin: attached mark on cells, marking them for destruction
Proteases: degrade proteins by breaking peptide bonds, converting protein into its amino acids
Proteasome: macromolecular machine that degrades proteins marked with ubiquitin
Ubiquitin-Proteasome Pathway
large cylindrical complex that proteins enter at one end and exit at other as amino acids or peptide fragments
marked by Ubiquitin
loaded into proteasome
degraded by proteases
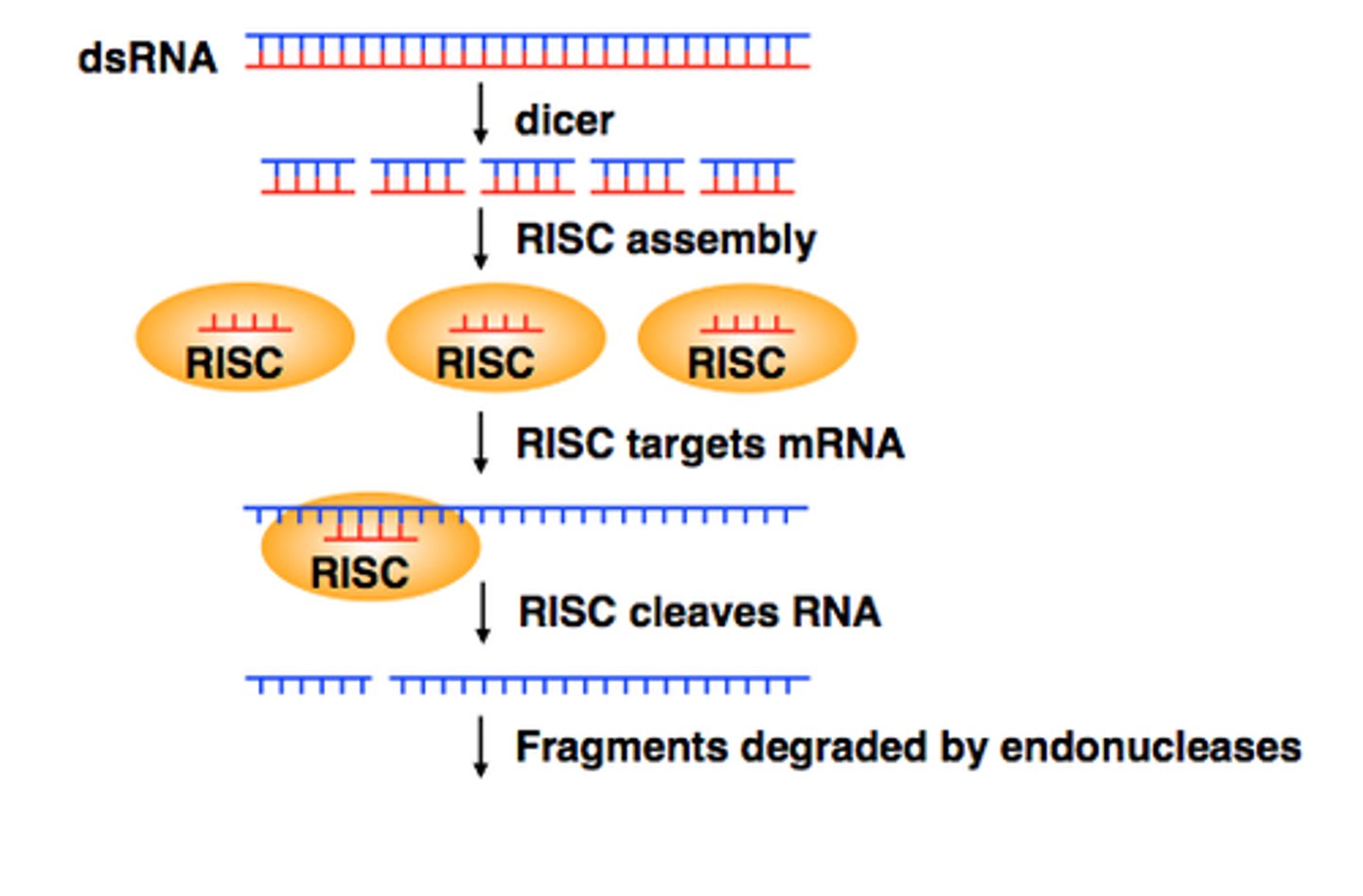
RNA-induced Silencing Complex (RISC)
complex of protein & RNA that can affect gene expression by inhibiting translation by degrading mRNAs
double sided pre-miRNA is cut with Dicer
strands of miRNA are loaded into RISC protein
bind to complementary mRNA
binding prevents transcription of that sequence