Biotechnology and Microscopy
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
48 Terms
Define optical microscopy
AKA light microscopy
Uses visible light and optical lenses to magnify and view a sample
Types
Fluorescent and compound
Define electron microscopy
Uses a focuses beam of electrons to magnify and view a sample
Larger and more expensive, but also more magnified
Types
Scanning electron microscopy (SEM)
Transmission Electron Microscopy (TEM)
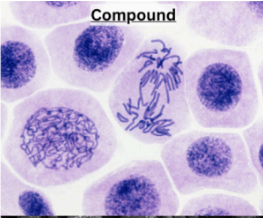
Describe compound microscopy
Visible light is focused on a thin slice of the sample
Produces 2D image
Uses
cells, tissues, organisms
staining used
Disadvantage
Can’t view living cells (stain kills)
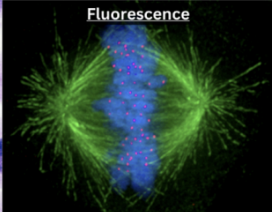
Describe fluorescence microscopy
Fluorescent marker to tag certain structures
Assist in visually locating protein expression within a cell
Uses
Thin slices of living samples
Can look at protein expression and specific parts of cell (chromosomes during mitosis)
Describe scanning electron microscopy (SEM)
Produces a 3D image of a sample’s surface
Sample must first be dehydrated and coated before viewing
Uses
High resolution with surface level detail (texture, shape, etc)
Ideal for viewing external surfaces of cells, tissues, and molecules

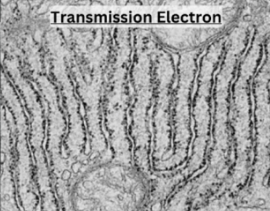
Describe Transmission Electron Microscopy (TEM)
Electron beam passes through a very thin section of sample
Produces a high magnification 2D image
Uses
Allows for high resolution viewing of internal structures
Ideal for viewing internal structure of cells, tissues, and organelles
High magnification
Disadvantages
Kills cells
Describe cell fractionation - process
Homogenization
Cell broken apart
Cellular homogenate (cell contents without membrane)
Low speed centrifugation
Creates dense pellet layer of nuclei
Scrape out pellet layer to study nuclei
Medium speed centrifugation
Remaining homogenate poured out and spun again
Mitochondria and chloroplast
High speed centrifugation
Process repeats
Leaving smallest components
Ribosomes and viruses
Describe vertical gene transfer
Transfer of genes from one generation to the next
Examples
Sexual/asexual reproduction
Mitosis
Describe horizontal gene transfer
Transfer of genes between different organisms
Three types
Conjugation
Transfer of DNA via a bridge
Between bacteria via a pilus
Transduction
DNA introduced into genome via a virus
Transformation
Absorb DNA from surrounding and incorporate into genome
Occurs via heatshocking and electroporating
Describe artificial recombinant technology
Use restriction enzymes to cut ip specific segments of DNA
Restriction enzyme cut at sequence-specific sites (recognition sites) —> palindromic sequences
Restriction enzymes produce:
Sticky ends - overhands of nucleotides (mostly used)
Blunt ends (no overhanG)
Describe the importance of restriction enzymes
Sticky ends allows fro new DNA pieces that are cut with the same restriction enzyme to bind
This creates a DNA molecule from multiple sources
Describe restriction mapping
Map of restriction enzyme cut-sites within a sequence of DNA
Useful to know where to cut DNA and what relevant sites are near by
Describe restriction fragment polymorphism (RFLPs)
Location of restruction site son human DNA will vary between individuals
DNA fingerprinting
Using RFLPs to link an individual to their own DNA in crime scenes or paternity
Similar fragments will appear
Describe single nucleotide polymorphims (SNPs)
Differences in human genome
May be found near disease associated alleles —> genetic markers
Describe gel electrophoresis
Used to separate DNA/RNA/proteins based on charge and size
Samples loaded at the top of the cell next to negative electrode
Moves to positive end
Separated based on charge and size
Smaller = further
After —> sequenced or probed to identify location of specific sequence
Probe —> radioactively labelled single strand nuclei acid used to tag a specific sequence
Proteins
Have strong folding structure and can be negative or positive
Needs to be treated with SDS to denature and make it into a linear chain
Also adds a negative charge coding
Describe nucleic acid hybridization
DNA or RNA form base pairs with complementary nucleic acids on a different strand
Used in DNA probing and in-situ hybridization
Describe DNA probing
DNA probe
Denture DNA probe into two single stranded pieces
If the DNA has the complementary piece, the DNA probe will bind to it
Use detectable label to see if a gene is present
Describe nucleic acid hybridization - in-situ hybridization
Tests the expression of a specific mRNA using a nucleic acid probe (DNA or RNA)
Probe labelled with a fluorescent dye
Probe hybridized with mRNA of interest
Fluorescent tag allows us to see the mRNA in place on the intact organisms
Can be visualized within tissues or small embryos
Define DNA sequencing
Used to determine the # of base pairs in a DNA or RNA molecule and their sequence
Early method
Dideoxy chain termination
Current
Next generation sequencing
Describe dideoxy chain termination sequencing
Denature DNA to separate strands
Single-stranded DNA is mixed with a primer
Primer provides 3’ OH necessary for DNA polymerase to begin DNA synthesis
Samples is incubated
DNA polymerase
dNTPs
ddNTPs (fluorescently tagged)
Lack 3’ OH —> cannot form phosphodiester bond
Deemed as replication terminating nucleotides
DNA polymerase adds until the terminating ddNTPs
Occurs many times over and we are synthesizing new strands
End up with strands of every possible lengths (different nucleotides have different fluorescent colour)
Separated via gel electrophoresis from shortest to longest strand
Each nucleotide is labelled and ordered digitally via a computer
Describe reverse transcriptase
Enzyme used to synthesize DNA molecule off an mRNA template
To create complementary (cDNA)
Since the template is RNA, there are no introns
Some viruses (HIV, Hepatitis B) use reverse transcriptase to replicate their genome and proliferate
Why make cDNA?
Required to create recombinant DNA in bacteria
Foreign DNA introduced into the bacteria cannot contain introns
Prokaryotic RNA does not contain introns, so they have no mechanisms in place to remove them
Allows for gene to be efficiently transcribe and translated
cDNA is much more stable and long-lasting compared to RNA
Describe the basics of polymerase chain reaction (PCR)
Important technique for the amplification of DNA
Necessary ingredients
Nucleotides
Primers
Heat-resistant polymerase (Taq polymerase)
Describe the steps of PCR
Denature
Separate into 2 strands via high temperatures
Requires heat resistant polymerase
Annealing
As temperature cools down, primers can attach to individual strands
DNA polymerase can only CONTINUE a strand, not make a new one
The primer allows for an attachment point
Elongation
The temperature is raised, heat resistant polymerase synthesizes complimentary strands
Keep in mind
Using a prokaryotic polymerase o human DNA still produces human DNA
Prokaryotic polymerase is more stable under heat
Occurs to both strands at the same time
PCR is run many times —> exponentially increase
Describe DNA microarray assay
Monitor the expression of large groups of genes across genome
See which genes are transcribe in different tissues or at different stages of development
Describe the steps of a DNA microarray assay
In a bunch of wells, there are short sequences of DNA to see which genes are actively being
Load cDNA into a pipette and add to the wells (fluorescently labelled)
No gene present = no hybridization = will not express any fluorescent signals
Analysis
Red —> in cancer cells only
Green —> normal cells only
Yellow —> both present
Grey —> not present
Used for:
Normal vs. cancer cells
Different cell types (nerve vs. macrophages)
Describe blotting techniques
Allow for the indentification of target fragments of DNA, RNA, or proteins
Types
Southern: DNA
Northern: RNA
Western: Proteins
Remember via SNOW DROP
To find if a particular gene sequence is presence
Describe the steps of southern blotting
Extract DNA with gene of interest via restriction enzymes
Separate DNA fragments by size via gel electrophoresis
Fragments transferred to nitrocellulose paper and blotting paper is stacked on top of it to create a sucking motion
Nitrocellulose paper exposed to labelled DNA probe
Allows DNA fragments to be visualized by hybridizing if present
Define immunofluorescent staining
Staining technique
Allows for the visual identification of proteins
List the steps of immunofluorescent staining
Addition of primary antibody to bind to a specific protein
Addition of secondary antibody which contains fluorescent tag and binds primary antibody
Visualization of protein of interest using microscopy, the fluorescent ga can be visually located to detect the protein of interest
Describe in-vivo mutagenesis
Helps determine the function of the gene by seeing what goes wrong without the functional copy of that gene
List the steps of in-vivo mutagenesis
Introduce a specific mutation into a gene to disrupt its functions
Observe for any phenotypic differences
Differences may be a function of a missing normal protein
Frequently used example
Knock out mice
In-vitro mutagenesis —> similar, but occurs OUTSIDE of a living organism (cells in a culture)
Describe genome annotation
Analyzing genomic sequences to identify the protein-coding regions and their functions
Utilizes computer databases to compare known sequences
Identifying functioning and non-functioning elements of a genome
Describe gene therapy
Introduction of genes into an afflicted individual for therapeutic purposes
Using a retroviral vector to insert genome material into chromosomal DNA
Non functional DNA segments have been replaced with functional ones
Describe transgenic animals
Animals which have a gene introduced from the genome of another individual
Transgenic mice —> implanted with a gene from a jellyfish that expresses a green fluorescent protein
Define genomic library
Collection of cloned DNA pieces from a genome
Library can be screened to locate a gene of interest
List the steps of creating a genomic library
Isolate genome of interest (extract DNA)
Cute genomic with restriction enzymes
Cut plasmid with same restriction enzymes (small, double stranded, circular DNA)
Ligate genes into plasmid
Insertion of plasmid into bacteria (via transformation)
Called making cell “compotent”
Using heat shock or electroporation
Allow bacteria to multiply to replicate genome
DNA isolation
Describe heat shock and electroporation
Electroporation
Brief electrical impulse
Creates temporary pores in plasma membrane
Heat shock
Temperature increased then rapidly cooled
Increases membrane permeability
How do we know if teh bacetria were transformed?
Plasmid used in genomic libraries contain antibiotic resistance genes
Antibody resistance test
Helps to verify which bacteria were successful in accepting the plasmid
Treat all bacteria with an antibiotic
Bacteria who didn’t survive indicate a lack of the antibiotic resistance gene
Unable to take up the plasmid
Bacteria who survive prove they are resistant to the antibiotic
Deemed ‘recombinant bacteria’ due to their successful uptake of the plasmid
List the steps of reproductive cloning
Isolate donor cell with nucleus
Isolate unfertilized enucleated egg from donor
Transplant nucleus into enucleated egg (electroporation)
Embryo formation
Transfer embryo into surrogate mother
Deliver baby clone
Complete clone to sheep that was the nucleus donor (somatic cell’s nucleus)
Describe Pasteurs Swan Neck Flask Experiment
Proved spontaneous life-generation was invalid
Life cannot be created from non-life
Describe Griffith’s experiment
Used 2 strains of bacteria that he injected into mice
Rough (R) strain —> lacked protective capsule and was therefore non-virulent (pathogenic)
Mic’s immune system killed the bad cells
Vector lives
Smooth (S) strain —> protective capsule shielding it from the immune system
Vector dies
Both R and S
Vector dies
The other plasma enters bacteria cells via transformations
Describe the Avery-McLeod-McCarty experiment
Separate addition of various digestive enzymes to the heat killed S’ bacteria
Digestive enzymes included: DNases, proteases, and lipases
Most of these mixtures had no effect
HOWEVER, when DNase was added to the heat-killed S’ bacteria, the R cells were not transformed
Any DNA in the tube was killed, which is needed for R’ bacteria to become virulent
R bacteria never gained ability to produce the protective capsule
Proteases + S’ bacteria would result in protein breakdown and unaffected DNA
Bacteria would still be transformed
Describe Hershey and Chase experiment
Showed that DNA, not proteins, was the genetic material of Phage T2 (a virus)
Virus that affects bacteria
Placed a radioactive label in DNA of the virus
Placed a radioactive label on sulfur in the protein of the virus
Sulfur is specific to proteins
None of radioactive labelled sulfur was identified in bacteria, but phosphorus was
Confirmed DNA is the genetic material of a virus
Describe Meselson and Stahl’s experiment
proved semiconservative replication model was the valid DNA model
1 Parent strand and 1 new strand
Grew E. Coli in medium with nucleotides
Bacteria transferred to medium with 14N
15N bacteria replicated in new medium
Replication continued for another round
Describe Gurdon’s Nuclear Transfer Experiment
Proved that fully differentiated cells do not lose their genetic information - and still retain full genome
Placed a nucleus from a differentiated frog cell into an enucleated egg cell —> gave rise to a new frog
Describe the purpose of the CRISPR/Cas9 System
System of bacterial defence against viral infections
Can search for and edit specific DNA sequences
Used as a form of biotechnology for gene editing
Cas9 protein: cuts DNA
Guide RNA (gRNA): recognizes the sequence to be cleaved
Describe the steps of the CRISPR/Cas9 System
Genome target sequence is identified
Complementary gRNA is synthesized to recognize target sequence
Cas9 and gRNA are inserted into target cell
Cas9-gRNA complex finds and cleaves DNA, allowing DNA to be removed or inserted