BIOL 3010 Exam 2 Vocab
1/118
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
119 Terms
tRNAs
a transfer RNA molecule that acts as an adaptor between mRNA and amino acid
charged by aminoacyl tRNA synthetases
contains anticodons complementary to the mRNA
attaches specific amino acids to the growing peptide chain
Wobble position
the anticodon of tRNAs participate in promiscuous base pairing at this position
can pair with standard or modified bases - accommodates degeneracy
less tRNAs than there are codons
RNA pol 1 & 3
transcribes rRNA to form ribosomes
(As opposed to pol2 that transcribes mRNA)
60S subunit
aka the large subunit of the ribosome
contains A and P sites, attaches last
Joins the small subunit during initiation of translation to form 80S
40S subunit
aka the small subunit of the ribosome
binds initiation factors that facilitate scanning of mRNAs and initiation of protein synthesis.
80S subunit
the assembly of small and large subunits together
responsible for protein synthesis
Initiation of translation
the small subunit of ribosome starts scanning down the length of mRNA to locate a start codon
when found, a charged tRNA interacts with a small subunit and allows assembly of the complete ribosome
methionine tRNA starts in the P site, and second one comes in through the A site
Elongation of translation
ribosome moves toward 3’ end of mRNA, tRNAs go from a→ p → e sites and are split out as the polypeptide grows
Termination of translation
stop codon contains a release factor that enters the A site and makes the ribosome dissociate/ fall off the mRNA
Kozak sequence
a preferred sequence surrounding the start codon that makes it more likely that the small subunit will recognize it and attach there
eukaryotic initiation factors (eIFs)
involved in the initiation of translation
-Associates with the 40S ribosome subunit, mRNA, and the 60S subunit to help stabilize and assemble the ribosome around the start codon
PABC
associated with poly (A) tail - this is why this aspect is involved in translational initiation
associates with mRNA, and eIFs to create looping where the 3’ end is close to the 5’ end
helps ribosome attach
Diamond-Blackfan Anemia
mutations in the small or large subunit create a stoichiometric imbalance between the units
results in depletion of mature ribosome → less efficient translation
especially crucial for stem blood cells to be efficient, but they are unable to replenish themselves
Treacher-Colin Syndrome
mandibulofacial dystosis that creates problems with swallowing, breathing, etc.
heterozygotes are the ones impacted
mostly due to mutation in TCOF1
normally recruits RNA pol1 to nucleolus
detrimental because it can’t create ribosomal RNA
TCOF1
treacle ribosome biogenesis factor 1
responsible for ~93% of cases of treacher-colin syndrome
in the nucleolus, responsible for recruiting RNA pol1 and localizing it in the nucleus where ribosomes can be made
amino-acyl tRNA synthetases
“charge” tRNAs by attaching an amino acid group on the amino end
uORFs
upstream open reading frames that regulate translation
start codons that lack a Kozak sequence in 5’ UTR
regulates genes that code very potent proteins or important in development
slows down the ribosome and ultimately translation because the small subunit is scanning down the mRNA, but signaling is poor
Sonic Hedgehog Signaling Pathway
important in body patterns during development
have PTCH receptors for Shh protein
The amount of signaling is important for the identity of cell (more dorsal or more ventral fates)
have uORFs that reduce amount of protein made to tightly regulate how much signaling in pathway
PTCH Receptor
protein that is regulated in the Sonic Hedgehog signaling pathway
amount of receptors determines body patterning
why are uORFs important?
reduction of protein
very normal and important for regulation / differentiation
especially in genes encoding potent proteins or ones involved in development
fluorescent reporter in uORF experiments
to learn the role of uORF
placed varying numbers of uORF from PTCH in front of coding sequence of FOXA2 gene
less uORF → more translational efficiency (more fluorescence)
Nuclease degradation of mRNA
mRNA degraded by ribonucleases, especially poly(A) nuclease
if you have a shorter tail, you have less PABC (important for translation initiation!)
microRNAs
aka miRNA- non-coding guides that bind to mRNAs
regulate mRNA abundance
by either targeting for degradation or preventing translation
must be processed by factors like Drosha, dicer
the single-strand associated with miRNA-induced silencing complex (includes RISC and argonaute proteins)
if perfectly complementary → target for degradation
if not, loop forms → prevents translation
Drosha
important in cropping miRNA during processing
- removes hairpin structure and the stem
in the nucleus!
Dicer
removes hairpin structure from stem in miRNA processing
in the cytoplasm!
TRAMP
targets the degradation of defective mRNAs in the nucleus
non-stop decay
transcript lacks a stop codon
ribosome tries to translate into 3’ UTR
no-go decay
RNA has structure (Ex. hair pin) that prevents ribosome from progressing
non-sense mediated decay
introduction of premature termination codons
results in truncated proteins - lack critical domain on carboxy end
EJC protein complex
exon junctional protein complexes
found right before every exon-exon junction
required for quality control
normally, ribosome removes proteins as it translates
but with PTC→ EJC is left and that acts as a signal to target the protein for degradation
B^0 thalassemia
non-sense mediated decay found→ PTC cause transcript loss and reduced hemoglobin
PTC
premature termination codon associated with non-sense mediated decay
results in truncated protein
What causes Zika virus?
Zika infects the neural stem cells
nucleus is oddly-shaped- has tri-spindle like apparatus
chromosomes unable to segregate properly
makes daughter cells triploid and they die
this example proves how important normal replication is to development
G1
first part of the interphase
the gap before duplication
S phase
when DNA synthesis and chromosome duplication occurs
-part of interphase
G2
interphase, gap before mitosis but after replication
G0
when a cell leaves the cell cycle
Role of growth factor in RTK pathway
ligands that activate the receptor tyrosine pathway (important for cell-cycle re-entry)
Overview of receptor tyrosine kinase pathway
receptors dimerize
ligands are phosphorylated and bring receptors together where they transphosphorylate each other
enables phosphorylation cascade of downstream targets such as GEF, RAS, RAF, MEK
ultimately activates kinases that phosphorylate a transcription factor
this promotes transcription of downstream genes (ex. cyclin)
Cyclins/CDKs
important for initiation of S phase
CDKs are cyclin-dependent kinases which need cyclin to function
bounded together → phosphorylate 100s of target proteins at serine - threonine residues
enables specific steps of the cell cycle
Cyclin D
transcription is driven by RTK pathway
binds to CDK-4
phosphorylate retinoblastoma protein to make it detatch from E2F protein → initiates S phase
Cyclin E
transcription is driven by RTK pathway
binds to CDK-2 → phosphorylates Rb so it dissociates from E2F→ activation of DNA synthesis
Rb protein
first identified as a tumor suppressor, but its main job is within cell cycle
bind to E2F to inhibit its activity
once phosphorylated, allows E2F to separate and initiate DNA synthesis
E2F protein
protein necessary for activating DNA synthesis
must be released from Rb to initiate DNA synthesis (activates genes)
p53 transcription factor
prevents G1 → S phase transition by
1)inducing expression of CDK inhibitor, p21
2) initiating apoptosis if severe
3)inducing expression of DNA repair enzymes
mutations in p53 promotes cancer susceptibility
main point in the elephant example?
“Peto’s Paradox”
large-bodied organisms typically live longer, but they have more cells and more time to accumulate cancer
how are cancer levels the same as small-bodied organisms?
additional copies of tumor suppressor genes like p53 allow for more mutations without risk of cancer
Cyclin A
pairs with CDK2 during S phase
-promotes DNA synthesis
Semiconservative DNA replication
-proposed by watson and crick
one strand of the original DNA acts as a template for a new strand
with further replications, original DNA will be present but less % of all DNA
Conservative DNA replication
2 original strands stay together and the whole thing acts as a template for a whole new strand
Dispersive DNA replication
different segments are replicated and daughter DNA is a mix of new and old
what adds nucleotides in replication?
DNA polymerase
-primer must be present
Origins of replication
recognized by CpG islands and promoters
-places where DNA replication may begin
Pre-replication complex (Pre-RC)
assemble at potential origins of replication
include ORC, CDC 6, MCM proteins
“license” origins of replication for use
only some become active
MCM complex
act as helicase to open/unwind the double strand
CDC6
loading the helicase and primase to prepare for replication
RPA (Replication Protein A)
keeps the single-strand from annealing before it can be replicated
Pol alpha
adds primer needed for replication
PCNA
acts as a sliding clamp to make sure new strand anneals to the template in DNA replication
Pol delta and epsilon
carries out replication off of DNA template
ligase 1
required for lagging strand- joins together Okazaki fragments
leading strand
continuous synthesis of DNA as the MCM complex and primer are moving the same direction
(polymerase chases replication fork)
lagging strand
discontinuous synthesis of DNA as MCM complex and polymerase are moving in opposite directions
replicated in portions - Okazaki fragments
polymerase must go back and replicate empty segments - requires ligase 1
“read-write” methylation
with chromatin remodeling - some parental histones remain in H3/H4 and some are newer- must adopt modifications
enzymes recognize particular histone modification in nearby nucleosomes and adopt the same methylation
persistence of heterochromatin marks
telomeres
occur at the chromosomes to prevent degradation of chromosomes after replication
repeats of TTAGGG
functions:
1) protect chromosome end from degradation
2) allows for own rejuvenation
3) pairing of homologous chromosomes during meiosis
G-overhang
150-300 bases of telomere that is single-stranded- must be protected with t/d loops!
T-loop
prevents the attack of G-overhang by nucleases
overhang folds over and invades other strand- pairing complementary bases
D-loop
result of G-overhang pairing with other strand
-aka displacement loop, smaller
Shelterin Complex
required for the formation of T-loop
telomere decorated with complex in G-overhang
Telomerase
active in the rejuvenation of telomeres
a ribonucleoprotein consisting of TERT enzyme and TERC
complementary to telomere repeats
uses RNA as a template for DNA
add sequences to G-overhang, then opposite strand filled in by DNA polymerase
telomerase reverse transcriptase (TERT)
enzyme that uses TERC RNA as template to synthesize complementary DNA
Diploid
describes cells carrying two matching sets of chromosomes; symbolized as 2x.
somatic cells of human body
Haploid
describes cells, organisms, or nuclei that contain one set of chromosome
gamete cells of humans
Sister-chromatids
identical copies of a chromosome joined together by centromere after replication
eventually split in anaphase
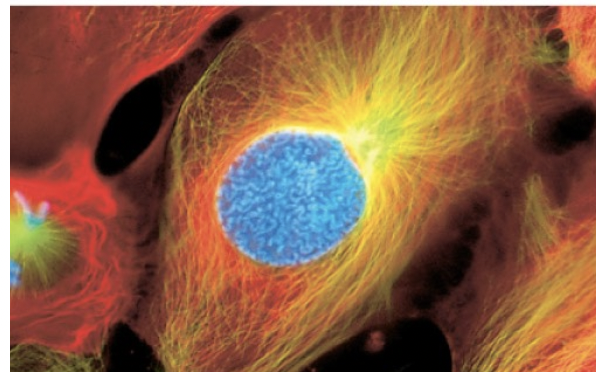
Prophase
-chromosomes condense and become visible
-centrosomes move toward poles
-nuclei begin to disappear
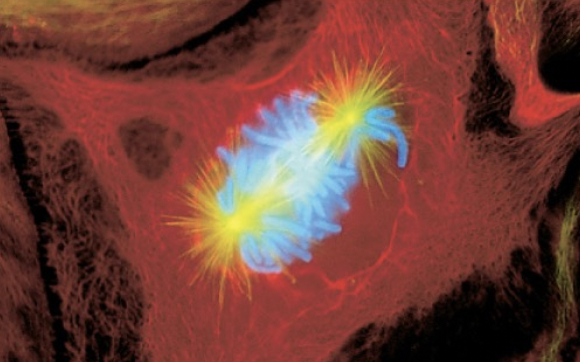
Prometaphase
-nuclear envelope breaks down
-centromeres invade the nucleus
-sister chromatids attach to microtubules
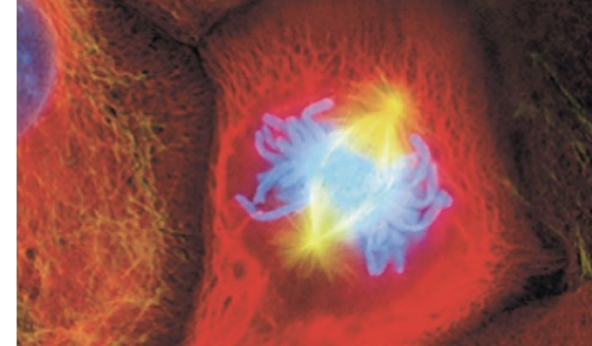
Metaphase
chromosomes align on the metaphase plate
sister chromatids face opposite poles
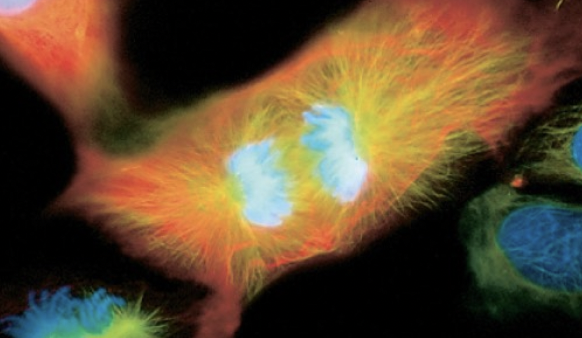
Anaphase
connections between centromeres severed, chromatids move to opposite poles
Telophase
nuclear membrane and nucleoli reform, spindle fibers disappear, chromosomes become tangled chromatin
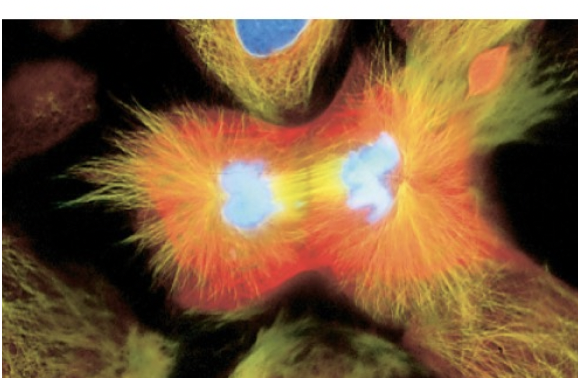
Cytokinesis
actual division of cytoplasm and rest of cell
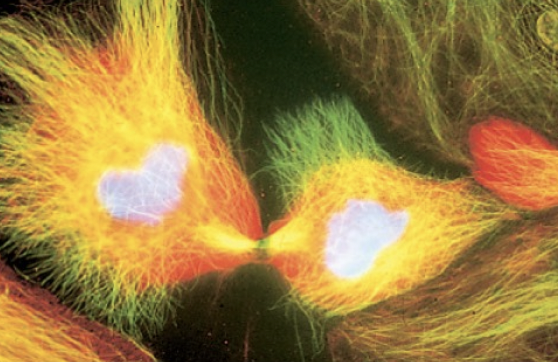
Cohesins
maintains contact between sister chromatids
form -quasi-ring around DNA
loaded in G1 phase but needs to be acetylated by ESCO1/2 during S phase to be stabilized
partially dissociates and only attached to centromere in prophase
ESCO1/ESCO2
acetylates cohesion during S phase to stabilize the complex
Separase
in anaphase, cohesion completely dissociates → done so by separas
Robert’s Syndrome
cohesion not stabilized by ESCO2
causes severe abnormalities of limbs and face as a result of slowed mitosis and damaged DNA
- sister chromatids not linked correctly
DNA packaging in mitosis
chromatin condensed into radial-loop scaffolds
condensin 1 and 2
make the loops that go out from center structure to condense the chromatin
topoisomerase lla
untangles and opens up the DNA molecule
-allow other molecule to pass through it
further condensing of chromatin
Centromeres
heterochromatin consisting of A-T-rich DNA
replaces H3 with variant called CENP-A to assemble to kinetochore
kinetochore joins chromatid to spindle microtubules
meiosis overview
one replication but two divisions → haploid gametes
only in germ cells
increased genetic diversity through independent assortment and crossing over
law of segregation
homologous chromosomes separate at meiosis 1
expect 3:1 ratio when crossing heterozygotes
law of independent assortment
pairs of homologous chromosomes separate independently of one another
allows new combinations of alleles in gametes
crossing over
exchange of genetic material of 1 chromatid to non-sister chromatid
Zygotenes
during prophase 1, onset of pairing homologous chromosomes at synaptonemal junctions
synaptonemal junctions
joins homologous chromosomes during crossing over
pachytene
assemble of recombination nodules at sites where crossing over occurs
stage of prophase 1
diplotene
recombination between chromosomes at chiasmata
back-crossing
crossing a heterozygote with one of the parental genotypes
method used in fly example about recombinant phenotypes displaying crossing over
overview of crossing over
Spo11 cleaves phosphodiester bonds
exonucleases degrade ends to expose single-stranded tails
strand invasion of non-sister chromatid → heteroduplex
reciprocal 2nd strand invasion
branch migration lengthens heteroduplex region
resolutions of holliday junctions
Spo 11
cleaves phosphodiester bonds at the beginning of crossing over to induce a double-strand break of the chromatid
heteroduplex
regions between holliday junctions where two strands may differ
holliday junction
X between non-sister chromatids during crossing over