AAMC Section Bank Biology
1/152
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
153 Terms
Based on the information in the passage, which type of enzyme is most likely to suppress CBC?
A. Phosphorylase
B. Kinase
C. Phosphatase
D. Synthase
passage: Phosphorylation of LC20 is required to activate the myosin head, which binds to actin... this will later create a CBC
C!
phosphatase!!
since phosphorylation activates CBC, dephosphorylation would to the opposite.
enzyme definitions
phosphorylation:
kinase
phosphatase
synthase
lyase
isomerase
ligase
hydrolase
phosphorylation: add phosphate to inorganic phosphate
kinase: add phosphate group from ATP to substrate
phosphatase: dephosphorylase substrate, remove phosphate group
synthase: link to molecules together via catalysis
lyase: catalyze cleavage without adding H2O
isomerase: catalyze conversion of isomers
ligase: bind 2 large biomolecules
hydrolase: catalyze cleavage with H2O addition
in vivo
take place in a living organism
Which statement regarding VSM function and cytoskeletal dynamics is best supported by the data in Table 1?
VSM function and cytoskeletal dynamics
vasoconstriction = expect smaller diameter.
during actin polymerization, monomers of G-actin polymerize into F actin.
Compared to control RMCA held at an internal pressure of 120 mmHg, application of latrunculin B to RMCA held at 120 mmHg will most likely result in:
decrease in F actin levels!
Passage states: actin depolymerization can be induced by drug latrunculin B.
while actin polymerization G actin is polymerized into F actin"
so adding latrunculin B will cause F actin -> G actin, therefore decreasing F actin levels.
According to the information in the passage, which statement best describes the function of the MR in response to stimulation of the sympathetic nervous system? The MR:
want to know the Mr function in sympathetic NS.
POE:
A- no, blood pressure is higher in aorta than in rest of body
B - yes, sympathetic NS cause increase BP
C - no, sympathetic NS causes vasoconstriction
D - no, blood flow is directed towards brain and skeletal muscles, NOT the organs.
most reasonable is B. due to high BP
The polymerization of which structural component was analyzed in the experiment described in the passage?
passage: polymerization of G actin.
actins are microfilaments therefore B is the answer
microtubules
help move things around (cilia, flagella, molecular motor)
microfilament
actin, makes up cytoskeleton
intermediate filament
maintain cell shape
thick filament
myosin
thin filament
actin
What is the likely structure of the amino acid found at position 19 of LC20?
passage states
"which activates an enzyme that phosphorylates amino acid residue 1- of the myosin light chain"
phosphorylates = OH!
so D (serine) is the correct answer
amino acid phosphorylation
THINK OF OH!
serine, threoine, tyrosine
Which table shows the expected body weight of WT and Gpcr43–/– mice fed a HFD while housed for 16 weeks in a conventional (CONV) condition, or a germ-free (GF) condition where the gut does not become colonized?
Passage: "microbial population...implicated the regulation of host metabolism, obesity & diabetes"
yes, you got this right, just look at figure 1 for the answer.
Since the independent variable is germ free (no gut microbiota) means that the WT would have more body weight because no metabolism can occur
In figure 1: we see that GPCR has lower overall body weight compared to WT. Therefore we expect the table should correspond to that under convention
for GF: as stated before, WT would have increased body weight. for GPCR, since it is able to regulate energy, we expect little to no change in the corresponding graph
Which amino acid is LEAST likely found in one of the transmembrane domains of GPCR43?
transmembrane protein = hydrophobic!
look for the amino acid that is polar
aspartic acid is the only one polar
hydrophobic amino acids
FAMILY VW
Compared to WT mice, which experimental group of mice is most likely to remain lean when fed a HFD?
if you look at figure 1, we see that without GPCR43, the body weight is significantly higher compared to GPCR43 present. Therefore we can conclude that GPCR leads to weight loss.
C
insulin: increase glucose -> fatter
acetate: decrease glucose -> leaner
also, we want to look at figure 1, where we see that if we compare GPCR (-/-) and WT, we see that GPCR (present in WT) is able to keep the body fat down. therefore we want GPCR to be overexpressed to remain "lean"
Which conclusion about glucose uptake is best supported by the data in Figure 2?
Figure 2.
insulin vs. acetate
insulin takes in glucose, acetate does the opposite.
if there is insulin only there is high glucose. both insulin and acetate its lower in WT. this means that insulin is suppressed by acetate!!
A
look at the figure carefully!
Insulin signaling results in the phosphorylation of the downstream target, Akt, which can be quantified by Western blot analysis. Which graphic shows the effect acetate administration will likely have on Akt phosphorylation levels in WAT and muscles of WT mice?
insulin + = akt high.
B is correct because in the present of insulin, Akt is high. but we also need to consider that acetate suppresses insulin.
bc insulin is suppressed, so is Akt here.
Compared to untreated WT mice, antibiotic treatment of WT mice is likely to result in:
Antibiotic treatment means less gut microbiota
look at figure 1, you see that WT on STD, the body weight is relatively low.
antibiotic treatment would do the opposite, and make him fatter. (B wrong)
increase adipocytes because if SCFA was removed by antibiotic treatment, then GPCR 43 would not be activated to reduce uptake into glucose. (C correct)
adipocytes
fat cells
Branched Amino Acids
LIV
Leucine, Isoleucine, Valine
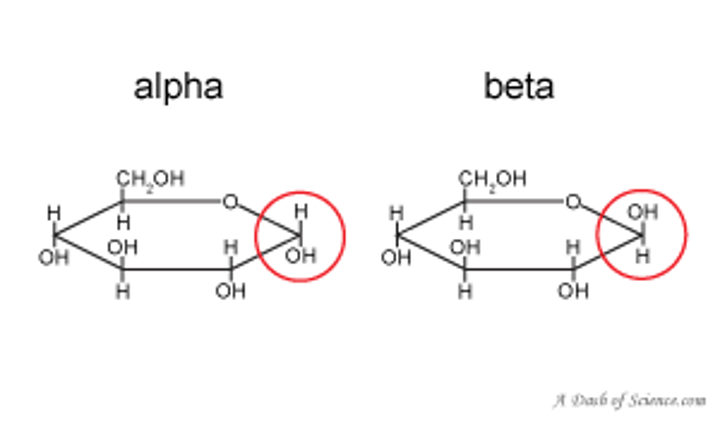
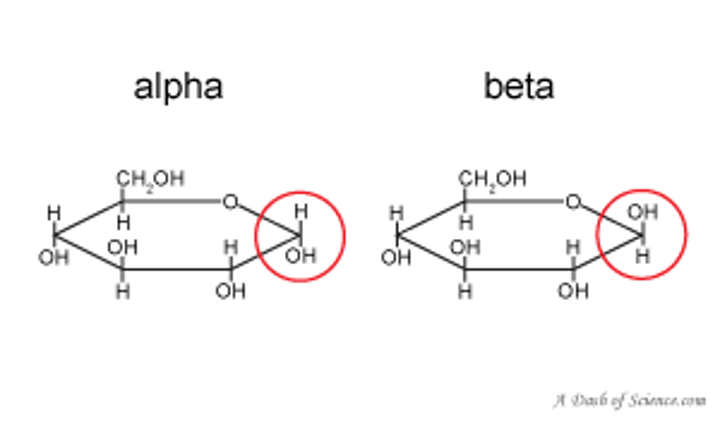
The stereochemical designators α and β distinguish between:
epimers at an anomeric carbon atom
alpha is a fish and fish goes down. (OH DOWN)
beta (OH UP)
when you think of alpha and beta for sterochemistry, think of anomers and the alpha and beta shit.
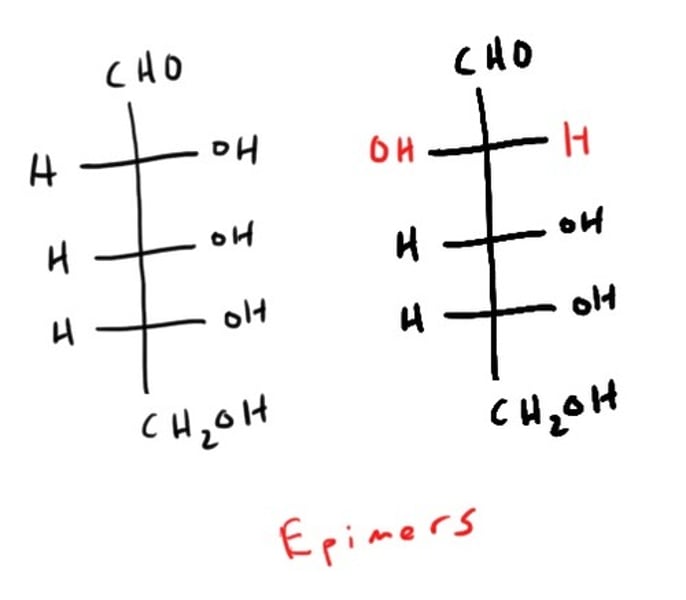
Epimer vs Anomers
epimers = different configuration in just 1 chiral carbon
anomers = different configuration in the chiral, anomeric carbon when the molecule is in cyclic form
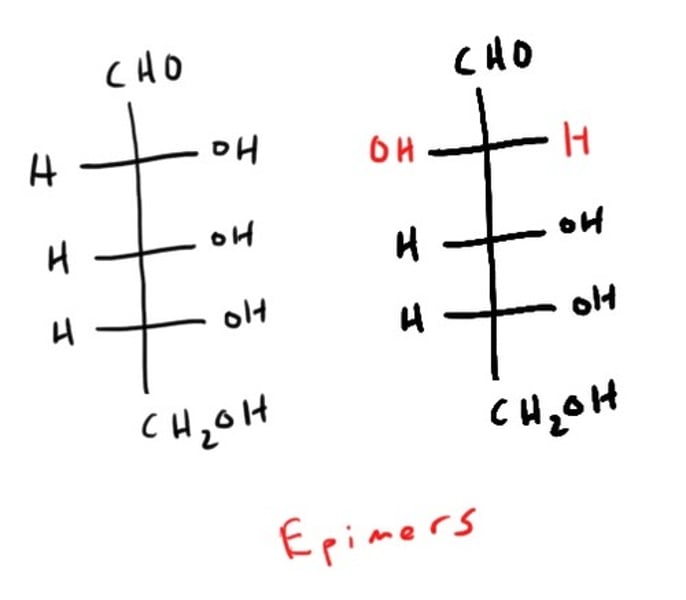
epimer
Diastereomers that differ at only one chiral center.
anomers
subtype of epimers that differ at the anomeric carbon
alpha = down
beta = up
Which ideal solution exhibits the greatest osmotic pressure?
easy question, you just dissociate the molecule into its ions then multiply
C:
0.2 M CaCl2
(0.2) x (3 ions) = 6.
(checked all the other answers already, and C has the greatest)
Which event is directly mediated by a ligand-gated ion channel?
Influx of Na+ across motor end plate resulting in the depolarization of the muscle fiber membrane!!
this is where voltage gated Na+ channels open for depolarization
A single point mutation in a gene results in a nonfunctional protein. Individuals heterozygous for this mutation were identified using a Southern blot. Which pair of wild-type (WT) and mutant alleles most likely contains the mutation?
Southern blot can only be useful if the mutation can either CREATE or ELIMINATE a restriction site (that are palindroms of 4 to 6 bp long) In this case, we see that AAGCTT is disrupted n A.
Southern blot uses restriction enzymes to cut DNA sequences and only cut at these sequences.
look at the answer choices and find the palindrome one.
What bond is cleaved by IN during the first reaction of integration?
passage "IN catalyzes cleavage of a GT dinucleotide from each 3' vDNA terminus"
it cleaves a guanine-thymine. (nucleotide) therefore it cleaves its phosphodiester bond here.
D is correct. (P-O)
A vDNA sequence encoding a protein is inserted into a host genome by IN. The protein is translated from the hypothetical mRNA sequence shown.
5-GGCAACUGACUA-3
Based on the passage, the segment of the original viral genome that encoded this protein had what nucleotide sequence?
5-GGCAACUGACUA-3
this has been translated.
viral DNA integrated into host cell genome by integrating would originate from a retrovirus. mRNA is then transcribed from retrovirus and can either used to synthesize proteins or used as the RNA genome for progeny virus
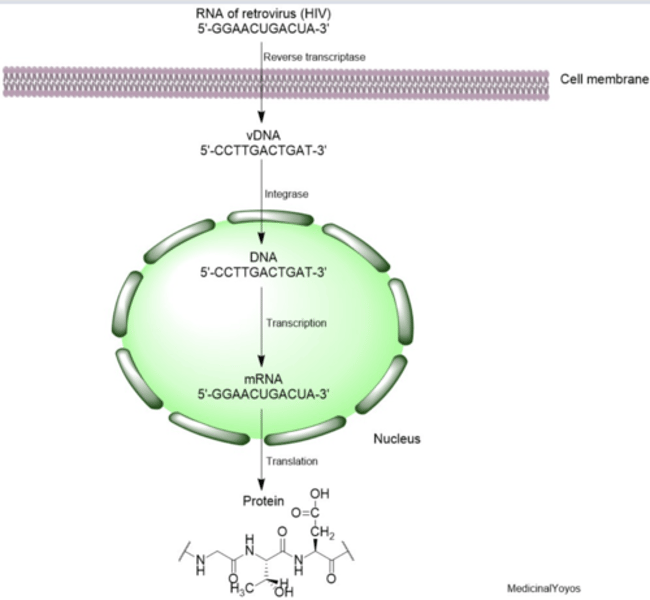
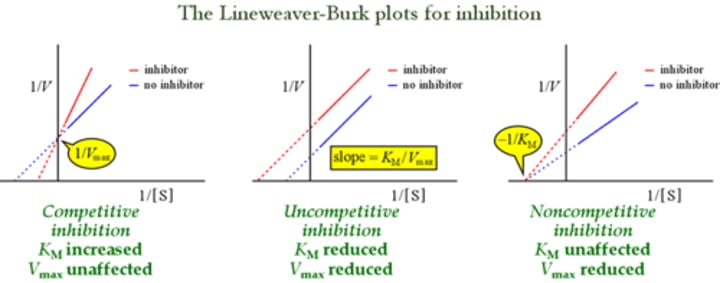
Which experiment would best provide data to support the mechanism by which an ODN inhibits IN activity?
"ODN are competitive inhibitors of vDNA"
remember the lineweaver plots
km: increase
vm: no affect.
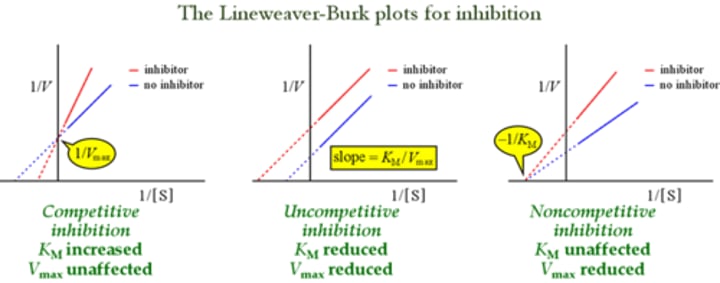
Lineweaver Plot Graphs
Which amino acid substitution for the conserved residue at position 64 is LEAST likely to affect the enzymatic function of IN?
D64 is aspartic acid.
you want a polar, negative charge amino acid in place.
so glutamic acid (E)
An inactive tetramer of IN is expected to have approximately what molecular weight?
Passage states:
288 residue protein
you also want a TETRAMER (question stem) 4 units
given that you know the molecular weight of protein (110kDa)
288 x 4 x 110
approx : 300 x 4 x 100 = 120000 Da = 120 kDa!
Average molecular weight of amino acid
110 kDa
To determine if a small molecule acts like a LEDGIN with respect to IN, a researcher plans to incubate purified IN both with and without the small molecule and then perform a Western blot to detect IN in each sample. Under which condition(s) should the gel electrophoresis step be performed?
I. Denaturing
II. Reducing
III. Native
Gel electrophoresis!
LEDGIN = inhibitors that bind IN and shift its oligomerization equilibrium toward inactive tertramer.
probably native, because you still want to know about its function. native will keep its natural/original function,
denaturing
disrupts interaction between monomers
reducing
disrupt disulfide bridges (created by cysteine)
native
does nothing to proteins, in its native state
SDS non reducing
eliminates quaternary structure by breaking hydrogen bond. It breaks into subunits (dimers into 2 and trimers into 3)
SDS reducing
eliminates quaternary & tertiary, broken disulfide bridge
SDS denaturing
eliminates quaternary, tertiary, and secondary.
only primary structure left.
Retinol uptake was measured in untransfected and STRA6-transfected cells in the presence of retinol bound to either RBP or bovine serum albumin (BSA). Uptake of retinol bound to RBP was also measured in cells that were cotransfected with a small inhibitor RNA (siRNA) targeting STRA6. Which graphic shows the expected result of this experiment?
Read the graph carefully. the binding protein STRA6 is on the X axis. so that means we are looking for the absence/presence of this molecule. In this case, untransfected is the control. The first two columns we expect the transfected column to be high, because there is an (-, absence) of the SiRNA, which is an inhibitor so we expect the levels to be high since there is no inhibitor. in the second column, BSA, we expect no uptake because BSA acts as a competitive inhibitor that prevents RBP from binding to retinol. This results in the levels to be low. in the last column, SiRNA is positive so there is inhibition, so we expect this level to be slightly lower than the first graph, but we still expect some uptake to happen.
Which statement is supported by the data shown in Figure 1?
Pay attention to the control and compare it with your results. Although LRAT, and CRBP lines are high, you still need to consider the STRA6 only line too. Since STRA6 line has a higher activity rate than the control, we still expect it to still produce activity. This means that STRA6 is not completely dependent on LRAT or CRBP for uptake. It can still uptake on its own, but a significantly lower rate BUT ITS STILL PRESENT
What is the dependent variable from the experiment shown in Figure 2?
Retinol fluorescence
Hill coefficient
A measure of cooperative interaction between protein subunits
hill coefficient = 0
no cooperativity
Hill coefficient greater than 1
positive cooperativity,
binding of one ligand facilitates binding of subsequent ligands at other sites
(increases affinity at remaining sites)
Which statement regarding STRA6-mediated retinol uptake is best supported by the passage?
Passage: "Retinal fluorescence, which is enhanced when retinol is bound to RBP, was measured over time"
higher retinal fluorescence -> lower reuptake
low retinal fluorescence -> higher reuptake
if you're confused, just compare it with the control, most usually you're trying to do the opposite to what the control is showing
since we are looking for reuptake, we are looking at the line that dips down the most.
We see that STRA6 + LRAT 0.5 holo dips down the most, meaning
"C" STRA6 becomes more dependent on LRAT for retinol release when the holo RBP to STRA6 molar ratio increases"
negative cooperativity
first binding event reduces affinity at remaining sites
From the data shown in Figure 2, which statement best explains the relationship between STRA6 and holo-RBP concentration in retinol release?
need some key information from the passage. we are trying to understand the retinol release by looking at the graph.
passage states that retinol flurescence is enhanced when retinol is bound to RBP
so for retinol release, we expect the opposite
(dec in fluorescene -> higher retinal release)
looking at the answer choices, C makes the most sense because:
STRA6 becomes more dependent on LRAT for retinol release (STRA6+LRAT line is lower than STRA6 only line).
when holo-RBP STRA6 ratio increases -> we see that there is less retinol release.
The organ in which the holo-RBP complex forms also functions to:
found in the liver.
it will detoxify!!
liver
produces bile, detoxify
secrete glucagon
alpha cells in the pancreas
produce HCl
parietal cells in the stomach
In a species of beetle, red body color is dominant to brown. Two red beetles are crossed and produce 31 red and 9 brown offspring (F1 generation). If two red F1 beetles are crossed, what is the probability that both red and brown beetles will appear in the F2 generation? (Note: Assume Mendelian inheritance patterns.)
4/9
What are the odds of crossing 2 hetereozygous F1 beetle?
(Rr x Rr) = 2/3
What are the odds of having both red & brown beetles in F2?
2/3 x 2/3 = 4/9
** Just break it up into simple punnet squares **
since it is asking for F2 generation, just break it up like that.
AND = multiply
OR = add
The diagram shows the size and position of the exons (numbered) and introns (lines) of a gene that codes for hypothetical Protein X, which can exist as two isoforms (either 16 or 17 amino acid residues long).
Which technique can be used to determine if a sample of cells expresses both isoforms?
1 amino acid consists of 3 residues. The isoforms NEED TO HAVE EXON 1 AND EXON 4. therefore the isoform 1 will have: exon 1, 2, 4, and isoform 2 will have exon 1, 3, 4. When you add the base pairs up and divide by 3. you will get either 16 or 17.
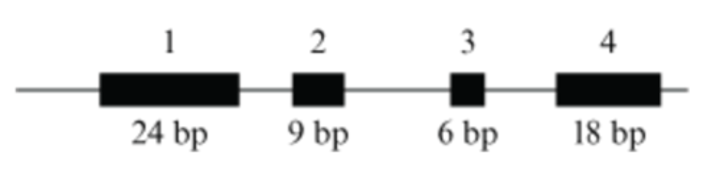
isoform
Related by different proteins. They can be generated by alternative splicing of a gene.
In Figure 1, which statement best explains the data trend of an increase followed by a decrease in bacterial cell count?
VP replicates inside the host cell before causing host cell lysis
*** all from the paragraph before the figure.
VP bacteria were injected onto the host cell.
Then, the sample was rinsed with ANTIBIOTIC that was effective against VP
** READ THE PARAGRAPH BEFORE FIGURE CAREFULLY, DRAW PIC AS NEEDED **
Figure 1 is measuring the lntracellular bacterial levels and we see that there is an increase then sharp decrease.
during incubation, cells grew (increase) the antibiotics made VP to lysis (decrease)
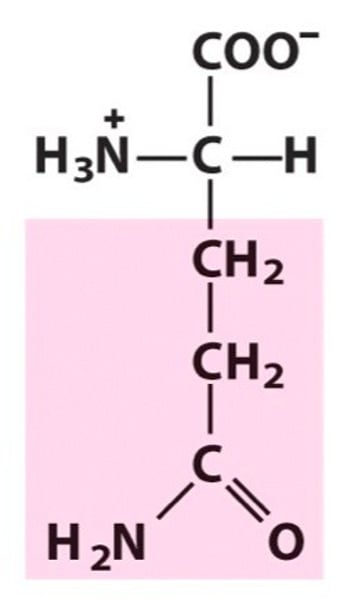
The amino acid residue at position 61 in Rac is most likely:
passage states: deamidation of the side chain of the residue position 61
glutamine!
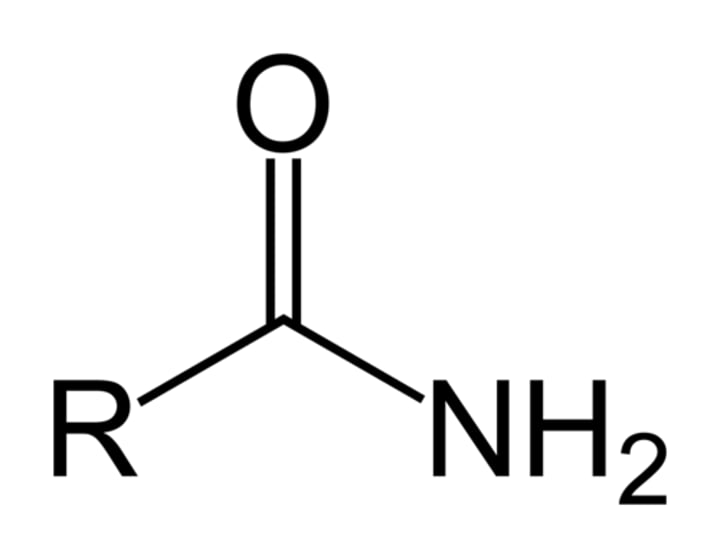
A Rac variant, in which the residue at position 61 was replaced with an alanine (Rac61A), was synthesized. Wild-type Rac and Rac61A were incubated separately with VopC. To obtain data to support that VopC modifies Rac at residue 61, the samples should be analyzed for the presence of which compound?
Passage: "VopC contains a catalytic domain that irreversibly activates host cell GTPase Rac thru deamidation of a side chain residue at position 61"
deamidation = amide group!
looking for group that has NH3
therefore B is the correct answer
Why was the total amount of Rac measured in Experiment 2?
Passage: "to confirm that VopC activates Rac, a mutant strain of VP lacking the VopC gene was created (VPvopC)"
keyword: activates
so we are thinking about transcription where things are activated on an mRNA. things will either be expressed or silence during this time.
D!
To determine if VopC affects Rac expression levels
Under normal conditions in the absence of VP infection, what is the structure of the molecule that binds and activates Rac?
Passage:
"VopC contains a catalytic domain that irreversibly activates host cell GTPase RAC thru the deamidation"
key word: GTPase!!
you're looking for GTP (guanine, so a carbonyl group, 2 ring)
The method used to visualize CREB327WT in Experiment 2 must include which step?
Passage: "CREB327 is phosphorylated by glycogen synthase kinase-3 (GSK-3) at serine 115. PKA phosphorylates CREB327 at serine 119."
Keyword: WT and PKA
pka we think of pH, WT we think control
"CREB327 WT cannot be phosphorylated at serine 115 or 119 were created"
dependent variable: "CREB327 phosphorylation by GSK-3 at serine 11.5.
B: establish a stable pH gradient in the gel before adding samples
Which conclusion about the phosphorylation of CREB327WT is supported by the data in figures 1 and 2?
think about the independent and dependent variable
independent: serine 119
dependent: phosphorylation
D: phosphorylation of CREB327WT by GSK 3 can only occur after phosphorylation with PKA
**just look at the figures and confer**
When designing variant CREB327119, which of the following residues was most likely chosen as the substitute for residue 119 in CREB327WT?
CREB327 119 has serine (polar, phosphorylation)
therefore we are looking for a control that doesn't have OH nor anything funky
Alanine!
Which statement explains how two forms of CREB can be generated in cells?
Passage: "Second messenger cAMP activates protein kinase A (PKA), which can then activate nuclear transcription factor cAMP response-element binding protein (CREB). CREB exists as two isoforms in cells, one being CREB327"
Keyword: second messenger, nuclear transcription factor, isoforms
we are dealing with transcription!!
isoforms: formed by splicing exons and introns; alternative splicing
B: CREB mRNA transcripts with different combination of exons are generated
isoform definition
Related by different proteins. They can be generated by alternative splicing of a gene.
Which statement about CREB327 phosphorylation is supported by the data presented in Table 1?
Passage: "Second messenger cAMP activates protein kinase A (PKA), which can then activate nuclear transcription factor cAMP response-element binding protein (CREB)."
second messenger activates PKA -> activates nuclear transcription factor
so, that tells us that PKA is the "iniatitor" to have other things activated
so, in fig 1 we see that PKA activates CREB, and it's even more activated with GSK-3 + PKA!
so: C - phosphorylation by PKA can partially activate CREB327
Under certain conditions, PKA and GSK-3 have been shown to autophosphorylate. The control group used in Experiment 2 (Lane 1) was designed to account for this possibility. Given this, the control group most likely contained
Passage: "CREB327WT was incubated with radiolabeled ATP and either PKA, GSK-3, or sequentially with PKA then GSK-3. Isoelectric focusing and autoradiography were used to detect phosphorylation (Figure 2)."
all the wells have ATP.
purpose of experiment: to see if PKA and GSK can phosphorylate on its own.
B: PKA and GSK 3 with ATP but with no CREB327 WT
Based on the passage, FSHRs are found on cells of which type(s) of tissue?
I. Connective
II. Epithelial
III. Nervous
Passage: "Follicle stimulating hormone (FSH) is a peptide hormone known to activate FSH G protein-coupled receptors (FSHRs) on ovarian cells. However, FSHRs have recently been found on osteoclasts, and their activation stimulates osteoclastogenesis and bone resorption."
Keyword: ovarian cells (epithelial) & bone (connective)
answer: connective and epithelial
Epithelial cells
skin cells that cover the outside of the body and line the internal surfaces of organs
example: digestive tract, bladder, air sacs, urinary system, reproductive system
Which conclusion is supported by the data presented in Figure 2?
passage:
FSH activates FSHR on ovarian cell & bones
estrogen inhibits these effects, but high FSH can lead to bone loss.
independent: specificity of FSH ab for FSH pep or FSH
dependent: intensity of fluoresence, increase absorbance for increased binding.
high absorbance = high binding
FSH has lower binding
A: The Kd for binding of FSHpep by FSH ab is lower than the Kd for binding of FSH by FSH ab
High Kd
low affinity for binding
Kd = dissociation constant
Which statement best explains why the absorbance levels for FSH differ from those for FSHpep?
Passage: An antibody (FSH-Ab) was then designed to bind the specific sequence of FSHpep. The antibody was also able to bind FSH.
FSH has lower absorbance level compared to FSH pep, means that there is lower binding.
There is no indication that there was cooperative or inhibition happing because passage says FSH ab was specific to FSHpep. therefore the only answer that makes sense is D
D: The tertiary structure of FSH limits FSH-ab binding interactions
Based on the FSHpep sequence, which amino acid substitution in the FSHR binding domain is most likely to have the greatest effect on reducing bone density loss in the presence of high levels of FSH?
Passage: "high levels of FSH, despite normal levels of estrogen, are associated with bone density loss."
FSHR activation increases bone density loss.
we need to decrease loss. (the opposite effect) therefore we need a substitution that disrupts binding
R8D changes from + to -
therefore it has most disruptive binding!
Which of the following reasons does NOT describe why FSHpep was included in the ELISA experiment?
- act as positive control to confirm that assay was functional
- to provide baseline against which to evaluate the affinity of FSH-Ab for FSH
- to generate data to support that the FSH-ab binds to receptor binding domain of FSH
Which statement describes a characteristic of FSH?
Passage: Follicle stimulating hormone (FSH) is a peptide hormone known to activate FSH G protein-coupled receptors (FSHRs) on ovarian cells.
Peptide hormone! they are water soluble therefore they do not need transport proteins to be in the bloodstream
steroid hormones need that.
so A
Peptide Hormone Characteristics
- water soluble (hydrophilic)
- binds to membrane bound receptor, triggers 2nd messenger system
Steroid Hormone Characteristics
- Fat soluble (hydrophobic)
- bind to carrier proteins for transport in bloodstream
- diffuses thru cell membrane & bind to receptors in cytosol & mitochondria
How do hydrophobic hormones get into the bloodstream?
using carrier proteins for transport
they are then diffused thru the cell membrane & bind to receptors in cytosol & mitochondria
this includes steroid hormones
Tyrosine Hormone Characteristics
- either water or fat soluble
- includes thyroid hormones (fat soluble)
Two gel electrophoresis analyses are performed on a sample of purified protein with unknown structure: SDS-PAGE (1 band appears) and SDS-PAGE under reducing conditions (2 bands appear). Which prediction about the protein is directly supported by these results? The protein:
contains multiple subunits!!
they were dissociated by the reducing agent as they has a disulfide bridge (caused by cysteine) between the subunits
Which type of interaction does NOT contribute to the stabilization of the tertiary structure of a protein?
Tertiary structures: (interactions bw R groups of amino acids)
- hydrophobic interactions
- van der waals
- disulfide bridge (cysteine)
- hydrogen bonding (OH, N)
- salt bridge (bw + and -)
not phophodiester bonds bc that's in DNA
Which type of inhibitor does NOT alter the KM/Vmax ratio of an enzyme?
Uncompetitive inhibitor!!
Km: reduced
Vm: reduced
just think about the lineweaver plot graphs!
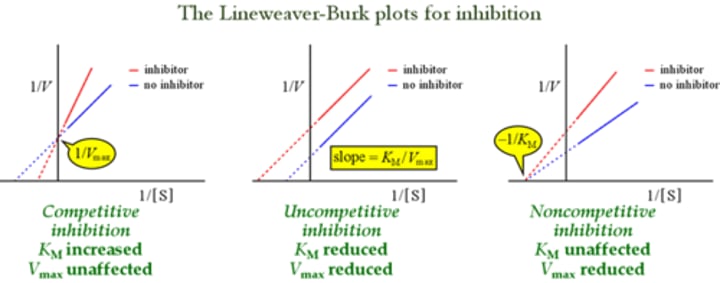
An enzyme is more effectively inhibited by uncompetitive inhibitors when:
I. the substrate concentration is decreased.
II. the substrate concentration is increased.
III. the inhibitor concentration is increased.
uncompetitive inhibitors
Km: reduced
Vm: reduced
uncompetitive inhibitors can only function with an enzyme-substrate complex. Therefore we want a higher substrate concentration
III yes because if there are more inhibitors, enzyme cannot bind
II yes because if there are too much substrates it won't function
Which peptide sequence is most likely found in a transmembrane helix of a protein?
transmembrane = hydrophobic!!
amino acids that are hydrophobic, nonpolar
F, A, M, I, L, Y, V, W
Ala - Ile - Phe - Val - Leu
In order to determine the effect of pRB on tumor growth, tumor cells containing no pRB (Rb–/–), basal levels of pRB (basal pRB), or additional induced amounts of pRB (induced pRB), were injected into mice. Tumor cell growth was measured at eleven days post graft. Which graphic shows the expected result of this experiment?
Passage: pRB may also pay a role in mitochondrial apoptosis
Easy question!!
just identify the independent, dependent, & control
control: no PRB (Rb-/-)
independent: basal pRB & induced pRB
dependent: tumor growth
C!
Which statement is best supported by the data shown in Figure 1?
Passage: "Cells with basal or induced pRB expression for 24 h were treated with the pro-apoptotic factor TNFα for 48 h"
"The mitochondrial apoptosis pathway involves the activation of the BAX and BAK proteins"
TNFα: increases apoptosis!
BAX and BAK proteins = facilitate apoptosis!
then look at fig 1 where Bak-/- and Bax -/-
the Bax - we see that there is no change
where Bak - we see that there is change.
This means that Bax is needed!!
Which experimental approach would be LEAST effective to determine the localization of pRB within a cell?
Passage:
"The retinoblastoma protein (pRB) has a primary role as a transcriptional coregulator of genes involved in several important cellular processes such as the cell cycle."
keyword: localization of pRB (wants to know where pRB is)
transcript is not needed to locate proteins. If you look through the answer choices you see that:
A tags the protein (makes sense).
C isolates their organelles (since pRB play a role in cellular processes, it makes sense)
D: pull down assay (measure protein-protein interactions; measure the affinity for protein), identification of interacting proteins using mass spect, this will help locate!
B: does not make sense, because the transcript of mRNA is not representative of where the protein is located, the mRNA will travel to the ribosomes for translation. ~ it will continue to move.
Rhodamine 123 is a fluorescent dye that binds to polarized membranes and is used to label mitochondria. A cell line expressing basal levels of pRB or which form of induced pRB would most likely exhibit the lowest level of rhodamine 123 staining?
Look at figure 2
"lowest level of rhodamine 123 staining"
apoptosis would effect the ability of rhodamine 123 to label the mitochondria from loss of membrane potential
Therefore higher the apoptosis rate -> lower level of rhodamine 123 staining
RB_SP has the highest percent apoptosis
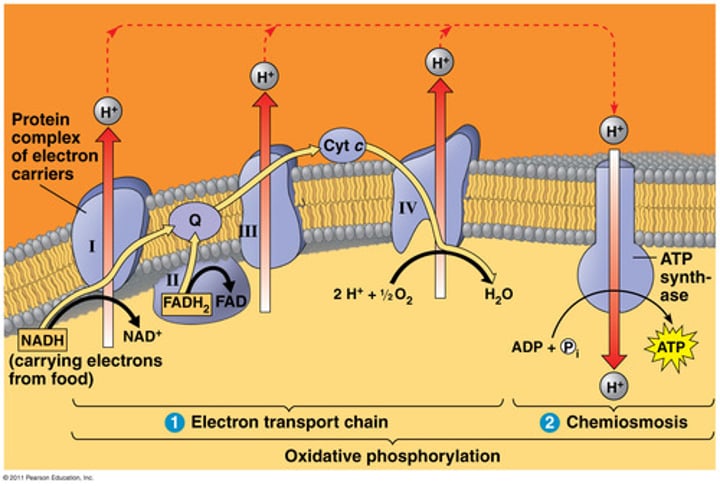
The caspase activator released during the mitochondrial apoptotic pathway primarily functions in which cellular process?
Passage: "mitochondrial apoptosis pathway... release of caspase activator cytochrome C"
electron transport chain!!
keyword: mitochondria = ETC, cytochrome C = ETC
cytochrome C
help carry electrons from one complex of the integral membrane protein to another
Cells were incubated with the indicated amounts of recombinant pRB only, or recombinant pRB in the presence of BAX and BAK. Following centrifugation, cell membranes were disrupted but organelles remained intact. Levels of cytochrome c in the supernatant (SN) and pellet fractions were analyzed by Western blot. Which graphic shows the expected result?
control: recombinant pRB only
independent: BAX & BAK presence in recombinant pRB
purpose: cell membrane disrupted but organelle intact
dependent: cytochrome C level & pellet fractions
we expect cytochrome C to have a presence in pellet.
we expect cytochrome C to have none in SN bc of the disruption of mitochondria (organelle), no longer have normal function
Based on the information in the passage, which protein domain of STAT3 is NOT predicted to play a role in its signaling?
A. nuclear localization domain
B. signal sequence domain
C. DNA binding domain
D. protein binding domain
A: "nuclear factor"
B: none
C: "nuclear factor"
D: "homodimer"
nuclear factor: requires nuclear localization domain for nuclear translocation & DNA binding domain
homodimer: protein made up of 2 identical polypeptide chains (protein binding!!)
Which amino acid substitution will most likely result in upregulation of leptin signaling?
Passage: "SOCS3 binds to Y985 within the LEPRb, thereby blocking recruitment of STAT3 to the LEPRb/JAK2 complex."
SOCS3 is blocking LEPRb from happening, therefore we need to have a substitution to prevent this
so changing Y985 to Y985F would be the best so SOCS3 can no longer bind and try to inhibit it.
Which mechanism restricts the expression of leptin to adipocytes? Only adipocytes contain:
Passage: "Leptin is encoded by the ob gene and is primarily expressed in adipocytes in response to feeding."
A: ob gene
B: promotor for ob gene
C: enhancer for ob gene
D: nuclear factors for ob gene
what is the only thing that can be found on adipocytes?
A: all cells contain the same DNA therefore the ob gene is most likely not exclusive to only adipocytes.
B & C: Promotors and enhancers are DNA sequences. They are also not unique to adipocytes.
D: nuclear factors are expressed on the ob gene, which are basically transcription factors. This makes sense because nuclear factors are the only elements that are different in cells.
nuclear factors = different in cells!!
Which amino acid substitution within the consensus-binding site for STAT3 is LEAST likely to interfere with STAT3 binding?
Passage: YXXQ
glutamine and asparagine are alike, as they are both polar. They differ as glutamine as an extra carbon in the chain.
Gln to Asn
Q to N