BIOL 325 CHROMOSOMAL STRUCTURE AND MUTATIONS STUDY GUIDE
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
56 Terms
what else can affect phenotypes other than gene sequences?
epigenetics
genes moving positions (translocation)
genes can be accidentally copied multiple times or deleted (epigenetic mutation)
alternation of control regions
alternations of chromosome structure or number
distinguish between the terms mutations (mitosis) and variants (meiosis) (written response question)
mutation is an inheritable change in genetic material. during mitosis, mutations can happen when there is an error in DNA replication or exposure to certain environmental conditions. there are three mutations: abnormal chromosomal number, abnormal chromosome structure, and small-scale, point mutation where frameshifts are the most detrimental.
variants are the cause of genetic syndrome (like down syndrome or marfans). during meiosis, recombination happens where the nonsister chromatids crossover resulting in variability.
what are the types of abnormal chromosomal number?
monosomy = one pair
polysomy = three or more pairs (down syndrome)
aneuploidy - has a wrong number of chromosomes
what is polymorphism?
two or more genetically determined (genetic issue mutations) that are proportionally represented types in the same population
what are some examples of balanced polymorphism?
if the polymorphism was bad for the human population, it would be eliminated by evolution
different versions of a gene that might be bad still exist because one bad gene and one good gene (heterozygotes) can be beneficial/better ability to survive
sickle cell allele —> one bad and one good = protect from malaria
some genes that encode juvenile diabetes protects miscarriage
what are some benign polymorphisms?
ABO blood groups
MHC (major histocompatibility complex)
eye color
thumb size
most variants are not good or bad, just different
what is the morphology of chromosomes?
eukaryotic chromosomes have linear DNA associated with a large amount of proteins
chromatin = DNA + histones condensed
chromosomes = compressed DNA and each offspring inherits one set of chromosomes from the mother and one set from the father
telomeres are at the end to protect them from fraying
sister chromosomes = identical copies of chromosomes
euchromatin = more exposed open DNA and heterochromatin = more compacted DNA
what will happen to the diameter of DNA during the cell cycle?
the diameter of DNA will change as it compacts down but will be uniform at each stage of compaction
what is the morphology of the DNA double helix during the cell cycle?
no proteins bound so it is 2nm diameter during interphase
what is the morphology of chromatin during the cell cycle?
chromatin is beads on a string
as the DNA is wrapped around the histone cores to create the nucleosome it will be 11 nm in diameter (G1, S, G2)
what is the morphology of chromatin in the nucleosomes during the cell cycle?
happens particularly in metaphase where most of the compaction happens (step 1)
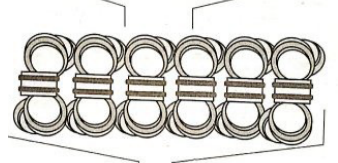
has a structure that looks like figure eight with something in the middle and is 30nm in diameter
what is the morphology of extended metaphase chromosomes during the cell cycle?
looks like wavelength and is 300 nm (step 2)

what is the morphology of condensed metaphase chromosomes during the cell cycle?
completely condensed chromosomes by the end of metaphase is 700 nm
what is mung bean exonuclease?
cuts DNA in linker regions between histones
can detect if histone placement is off because histones should be perfectly placed at the right location
explain how DNA fragmentation assays detect apoptosis (written response question)
DNA fragmentation assays can detect apoptosis through the DNA ladder formation that occurs when a cell is undergoing apoptosis. Apoptosis is often seen when run on an agarose gel. Before the cell goes through apoptosis, it will appear as one band on an agarose gel and when a cell that is undergoing apoptosis starts losing the chromatin “beads on a string” structure and the linkers will become visible and will get chopped by mung bean exonuclease.
how is X-activation detected?
it is painted with things that tag the Barr body specifically with a fluorochrome/fluorophore
probed with mRNA called Xist RNA
small RNA coats the Barr body so you can visualize the silenced Barr body
understand these various structural changes: translocation, duplication, and deletion of chromosomal regions (written response question)
translocation is when a DNA segment from one chromosome moves to a different chromosome
duplication is when a DNA segment is copied one or more times resulting in an extra copy of the DNA segment
deletion is the removal of a region from a chromosome and if it is large enough, it can be seen
what happens in metaphase? what is cohesion? what is condensin?
in metaphase, histones tightly wrap chromosomes
cohesion helps the sister chromatids stay attached to each other during the cell cycle
condensin proteins helps relax/maintain the DNA in interphase
what happens in prophase 1? what is a chiasmata?
synapsis occurs
the nonsisters line up with each other and cross over and undergo recombination
occurs more than 90% of the time required for meiosis
sometimes recombination happens when it shouldn’t happen and it leads to chromosomal abnormalities that very frequently lead to death or cancer
chiasmata = when the two non-sister chromosomes get into contact with each other exchanging information
what are centromeres?
it is always probed
holds the two sisters to each other
location of centromere varies in each chromosome
centromeres are very repetitive sequences and are very rich in AT base pairs
can be visualized in karyotyping with centromere-specific probes
what is metacentric?
centromere is right in the middle
what is submetacentric?
centromere is off center
what is acrocentric?
centromere is really close to the ends
how do you read a chromosome? what is an example?
p = short arm and q = long arm
first number = region on the chromosome
region one is close to the centromere
second number = band on the chromosome
band close to the centromere is one
third number = sub-band
an example = cystic fibrosis transmembrane receptor
7 q 3 1 . 2 and is read like 7th chromosome, long arm, region 3, band 1, sub-band 2
how do you visualize metaphase chromosomes with banding?
patients cells are grown and treated in tissue culture so to stimulate cell division you would use phytohemagglutinin (PHA)
after stimulating cells to divide and it is in metaphase, use colcemid which arrests the cells in metaphase which is the best in karyotyping
fixatives are added to stick something on a slide or stop it to preserve it so it can be analyzed and stained
3:1 methanol:acetic acid is added to fix metaphase chromosomes for staining
what are three banding stains? what are fluorescent probes used for?
giemsa = bands are stained black which are the AT rich sequences
cheap and easy but only one color
reverse = opposite of giemsa —> bands are white
centromere-specific stains = for metaphase chromosomes and only used if you have the money and needs for it
fluorescent probes are used to color the chromosomes differently and are not just one color
what is quinicrine?
binds to AT rich areas like giemsa
the dark parts in the quinicrine are DNA insensitive meaning that DNase cannot cut it, LINE rich, ALU poor, condense early, replicate late
if you stain it quickly and then take it off, you can find the area in the chromosome that condensed first
if an area condenses first early consistently then there is likely not many genes in that area meaning that it is not always critical to analyze that region
the light bands are the opposite of the dark bands
define karyotyping and what it can be used to detect (written response question)
karyotyping is a test to check for genetic abnormalities in chromosomes. it can be used to detect an abnormal number of chromosomes like aneuploidy. it can also be used to detect structural changes in chromosomes like translocations, deletions, and duplications. karyotyping is often used in pregnant women over the age of 36 because genetic abnormalities are common in them.
what is aneuploidy? what is an example?
a copy of an extra chromosome or missing a chromosome
aneuploidy in the sex chromosomes is the most common to check for
ex. 48, XXXX
human species and most animals have evolved to not care of how many sex chromosomes they have
could have 0, 1, or 2 and be ok
what is trisomy and what is an example?
trisomy is a type of aneuploidy and is common
condition where there is an extra copy of a chromosome
ex. trisomy 21 which is down syndrome
47, XX, +21
why don’t smaller chromosomes don’t always separate correctly? why are bigger chromosomes uncommon for aneuploidy?
it is hard for the meiotic spindle to grab onto small chromosomes
bigger chromosomes = very uncommon for aneuploidy because they are easier to separate and if there is a messed up 3rd chromosome then it is not compatible with life
what is special karyotyping? what is it able to define?
developed to unambiguously display and identify all chromosomes in a different color without prior knowledge of any abnormalities
five spectrally distinct dyes are used singly or in combination to create a cocktail of probes, creating unique spectral signature for each chromosome
able to define translocations and complex rearrangements; cannot detect intrachromosomal rearrangements such as duplications, very small deletions, or small paracentric inversions
what types of stains are used to analyze chromosomes and karyotypes? (written response question)
banding can be used to analyze chromosomes and karyotypes. giemsa banding is a common, cheap, and easy stain that stains the AT rich sequences in a chromosome and shows unique banding patterns. fluorescent probes can be used as well for the analysis of chromosomes and karyotypes. quinacrine is a common fluorescent probe that binds to AT-rich areas and the dark areas are DNase insensitive, LINE rich and ALU poor, and can tell if it condensed early or replicated late.
what are the 7 chromosomal structure abnormalities? describe them
translocation = a DNA segment from one chromosome moves to a different chromosome (1 —> 3)
deletions = removal of a region from a chromosome (if it is a large segment —> can see)
inversion = happens within the same chromosome and it is when something from the q arm moves down and inverts upside down into the opposite p arm
isochromosome = not common but catastrophic when it happens, when it is replication the q segment is repeated and the p is lost or vice versa
derivative chromosome = created by the translocation and it is when a chromosome becomes a part of another chromosome
insertion = a part from a different chromosome gets inserted/unique information gets inserted
ring chromosome = is rare and occurs when the telomeres get lost and goes into a ring formation to protect itself
what is a philadelphia chromosome?
first discovered and described in 1959
first genetic defect to be linked with a specific human cancer
it is a reciprocal translocation where two chromosomes both exchange information to each other
found in 30% of ALL
presence of translocation = diagnosis of CML and 98% of CML is positive for BCR-ABL1
what is the chromosome mapping of the philadelphia chromosome? what does it contain?
t(9,22)(q34:q11) —> translocation between 9 and 22; q34 of 9 and q11 of 22
contains a fusion gene called BCR-ABL 1
BCR = makes protein product
ABL = tyrosine kinase
the ABL1 tyrosine kinase activity of BCR-ABL1 is elevated compared to a wild type ABL1
this translocation messed up DNA repair because ABL1 is used for DNA repair
expression is changed by putting it next to BCR
understand the basics of FISH (metaphase or interphase) technology (written response question)
the basics of FISH for interphase technology is that it can pick up big scale things like normal diploid signal, trisomy or insertion, and or monosomy or deletion. two signals are used to probe in a cell. FISH for interphase can detect aneuploidy but cannot detect all abnormalities. it can also detect common translocations which can result in cancer by using a fusion probe.
what is the bait in FISH?
fluorescent nucleotide probe to a DNA sequence
what is the prey in FISH?
total gDNA in chromosomes from blood cells or cell lines on glass slides
what is the fishing pole in FISH?
fluorescent microscope
what are the steps for fish? what is hybridized to the immobilized chromosomes?
on the slides are fixed cells (biopsy)
the tissue is fixed on the slide by using methanol:acetic acid
permeabilize it because the membrane is in the way
denature nucleic acid
add fluorescent probe
wash
counterstain
put a slide cover on top and mount it on there so it can be evaluated
hybridization of complementary gene or region-specific fluorescent probes to immobilized chromosomes
what is direct labeling of probes?
uses nucleotides that have been directly modified to contain a fluorophore
what is indirect labeling of probes? what is hapten?
used more commonly
probes are labeled with modified nucleotides that contain a hapten
hapten is something that interacts with an antibody but does not induce an immune reaction in the body and a commercially created antibody is created to interact with the hapten
what is used in metaphase fish?
chromosome painting
spectral karyotyping
what are the uses for FISH for metaphase and interphase?
identification and characterization of numerical and structural chromosome abnormalities
detection of microscopically invisible deletions
detection of sub-telomeric aberrations
prenatal diagnosis of the common aneuploidies (interphase FISH)
what are the four FISH probes and describe them?
chromosome-specific centromere probes (CEP)
detect aneuploidy in interphase and metaphase
easy to keep making because the sequences of the centromeres are known
chromosome painting probes (WCP)
whole chromosomes or region
easy to paint whole chromosome because we are 99.9% identical
characterize chromosomal structure changes in metaphase cells
unique DNA sequence probes (LSI)
detect gene rearrangements, deletions, and amplifications
empiregenomics.com, abnova, vizgen, molecular.abbot
telomere-specific probes (TEL) abnova, PNABio
detect subtelomeric deletions and rearrangements
subtelomere probe would be right next to the repeated sequence of a telomere
in what situation where you would use telomere probes?
there are common disorders that lead to intellectual disabilities that are due to mutations or loss of subtelomere region
why is a WCP probe used? what is an example of a WCP probe being used for translocation of metaphase FISH?
used if you don’t know what is going on
ex. women has pain and the doctors didn’t know why so they painted her chromosomes and analyzed them
found a translocation of long arm 7 to short arm 5 which lead to premature ovarian failure
what is williams-beuren syndrome? what are the phenotypes for williams-beuren syndrome?
chromosome 7 is deleted, right arm
7 q 1 1 . 2 3
the lose of the band lead to loss of 25 genes
“happy baby”
phenotypes
cognitive challenges
outgoing personality
wide spaced eyes
flat nose
blood vessel problems
how can you predict the deletion of the 7th chromosome in williams-beuren? what is the hypothesis?
region of chromosome 7 has many repeats
repeats may lead to inversions (naturally) and inversions can cause deletions because the repetitive sequences can be skipped over and skip over important genes
hypothesis: are inversions more common in donating parents than general population?
how is interphase fish and fiber fish used in W-B parents?
used to see if inversion is common in parents of W-B parents
requires 3 color FISH
order genes on 7Q
what is fiber FISH? what are the steps to the technique?
overcomes FISH resolution limits with metaphase chromosomes
if you look at the metaphase chromosomes, you might miss things that are compacted like DNA because you can’t see them with probes so it does not use metaphase chromosomes
makes chromosomes stretch out into a long fiber on a slide
with the chromosomes being stretched out, you can look at all the DNA elements by fibering the chromosomes by improving resolution to 1,00 bp
not a tough technique
put DNA on a slide
add reagent
rxn happens that pulls the DNA and stretches it
what method was developed in 1993?
parra and wilde developed a method which uses a glass surface to capture DNA fibers directly from cells that were lysed
lysed cells with a detergent at one end of the glass slide and let DNA in solution stream hydrodynamically down
used fluorescent hybridization to map specific locations within the fibers
what is a real world application for fiber FISH?
found location and movement of IGT with fiber FISH
what is loss of heterogeneity?
loss of the heterozygous gene on the other chromosome and is not in banding
heterogenity can be picked up with banding
name a reason FISH might be used for molecular diagnostics (written response question)
one reason that FISH might be used for molecular diagnostics is to identify and characterize of numerical and structure chromosome abnormalities. FISH can be used to detect translocations in chromosomes. FISH can also be used to detect aneuploidy in metaphase or interphase chromosomes. FISH can also be used for prenatal testing.