Biochem 285 Week 5
1/22
Earn XP
Description and Tags
DNA Replication and Structure, Genome Organization, and Histone Modification
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
23 Terms
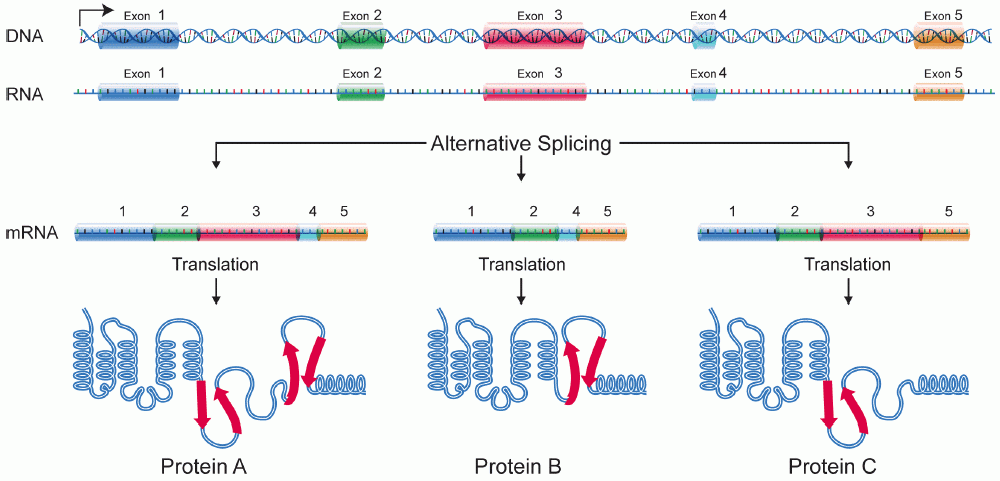
A single eukaryotic gene can lead to the production of many different proteins, why?
This is due to alternative splicing of mRNA transcripts. One gene can be transcribed for multiple proteins. Prokaryotic genes can’t do this because they lack introns and thus only code for one protein per mRNA transcript.
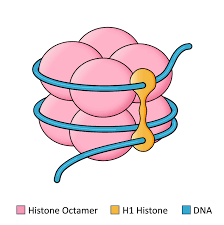
Where do chemical histone modifications occur?
At the N-terminal tail of histone proteins. There are 8 subunits of proteins around a nucleosome core, and they all have a N-terminus tail sticking out that can be modified.
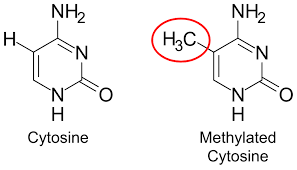
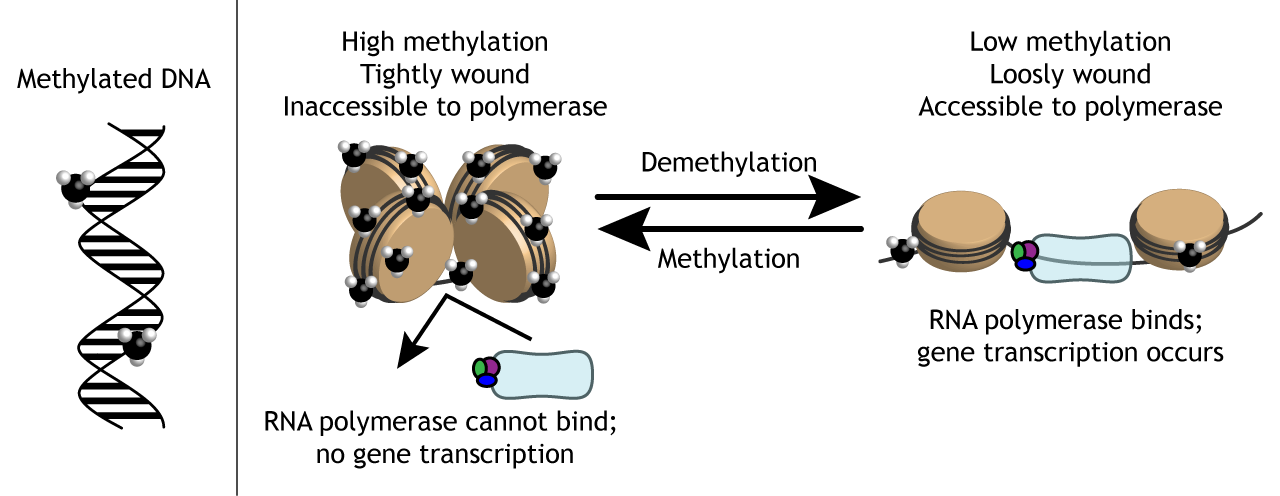
What is DNA methylation and where does it occur?
It is the process of HMT (enzyme) adding a methyl group to cytosines on CpG islands. CpG islands are located near the promoter region. This works to silence gene expression by blocking proteins related to transcription to bind. Methyl groups are uncharged. It competes with acetylation because they both modify lysine.
Are epigenetics heritable?
Yes, epigenetics are heritable but they are also affected by environmental conditions.
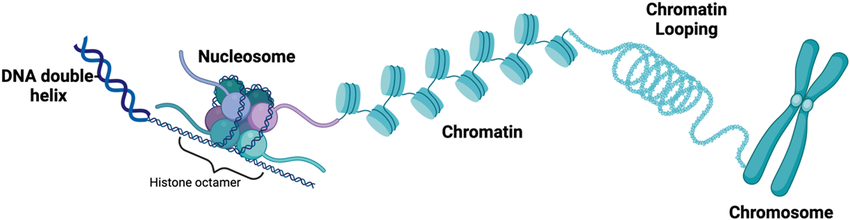
What are the components of a nucleosome? How do they regulate chromatin structure.
Nucleosomes are units of DNA packing in eukaryotes that have an octamer of histones and DNA wrapped around them (~1.5 times). They regulate chromatin structure with the N-terminal tails that can be chemically altered through modifications like phosphorylation, acetylation, or methylation. These modifications either cause the chromatin to avoid gene expression by tightening (heterochromatin) or activate gene expression by loosening (euchromatin).
Purines
Adenine and Guanine
Pyrimidine
Cytosine, Uracil, and Thymine
Histone Acetyltransferase (HAT)
Enzyme that transfers acetyl groups onto lysine. Lysine is positively charged and by adding a negatively charged acetyl group, it neutralizes the charge on lysine making it no longer attractive to DNA. This lack of electrostatic interaction allows for the chromatin to loosen and thereby increasing gene expression.
Leading Strand
The strand synthesized in the 5’ to 3’ direction utilizing one primer continuously.
Lagging Strand
The strand is synthesized in Okazaki fragments due to it being in the 3’ to 5’ direction. DNAP has trouble adding bases in this direction so it needs to do it in fragments. This causes the process to be slower than its counterpart strand.
What is a primer in DNA replication?
A primer is a short RNA sequence that initiates DNA synthesis. It is necessary because DNAP is a little bitch and can’t start adding dNTPs unless there’s an existing 3’ OH end. The primer gives something for DNAP to bind to initially and is set down by RNAP at first because RNAP is actually capable. The primer being RNA makes it easier to remove afterwards as RNA to DNA base pairings are easier to break than DNA to DNA base pairings
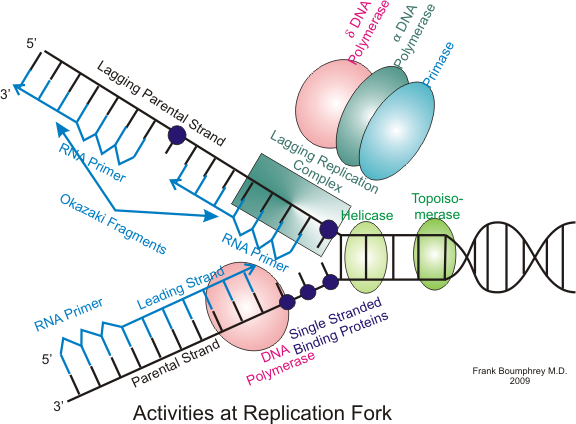
What is the end replication problem?
On the lagging strand, DNAP adds dNTPs in Okazaki fragments because of the strand’s unpreferable 3’ to 5’ direction. But remember how DNAP is a little bitch? Yeah, it needs an RNA primer for each of the Okazaki fragments and when each of those primers clock out for the day (leave the fragment), they leave a gap in that part of the new strand making for shorter and shorter telomeres. This is a problem. So cells have evolved to have telomerase come in and fix the issue by adding repetitive DNA sequences to the ends using its own RNA template so that a primer may come in.
Prokaryotic genome organization
Less DNA but more of it is coding sequence. Circular genomes. Not associated with introns.
Eukaryotic genome organization
More non-coding sequences and is associated with introns. Linear genome.
Chromatins
Linear DNA wound around histones and non-histone proteins.
Acetylation
The addition of an acetyl (-) group. Typically activates gene expression. It competes with methylation because they both modify lysine.
Phosphorylation
The addition of a phosphate (-) group. Can either activate or silence gene expression but is more associated with activation. It does this by encouraging repulsion between histones and DNA, by adding negative charge to modified amino acids. Typically occurs on Serine, Threonine, and Tyrosine because the process modifies the hydroxyl group present in them. Kinases add phosphate groups, phosphatases remove.
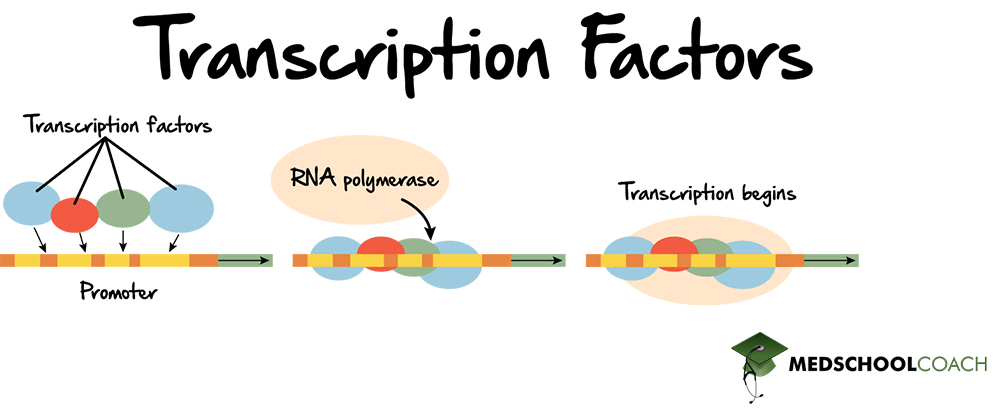
Transcription Factors
DNA binding proteins that promote gene expression by recruiting/helping RNAP to bind promoters.
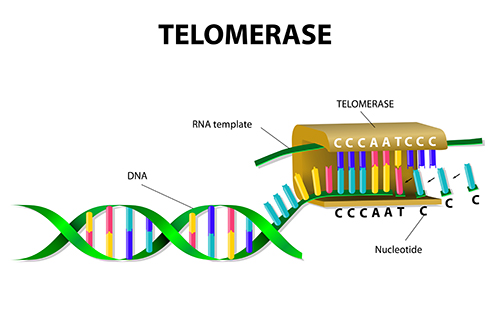
Telomerase
Ribonucleic protein (has both RNA and DNA component) that adds telomeric repeat sequences to the 3’ end of DNA that served as the template in the last round of replication. Contributes to cellular life span and helps regulate cellular senescence.
Boundary Elements
These things exist to prevent excessive histone modifications as you usually only want part of the genome to be expressed. An example who be a group of methylated histones because you cannot acetylate something that already has a methyl group on it and vice versa.
Writer proteins
Enzymes that catalyze the addition of chemical groups (modifications) by recognizing specific amino acids or DNA sites. Histone Acetyltransferase (HAT), Histone Methyltransferase (HMT), and Kinases
Eraser proteins
Enzymes that reverse previous epigenetic modifications by recognizing specific modified sites. Histone Deacetylase (HDAC) and Histone Demethylase (HDM).
Reader proteins
Proteins that bind to specific modifications on the histone and serve as a binding site for other proteins which may be involved in repair, gene expression, or silencing.