L5.2 – Histone modification
1/64
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
65 Terms
ER - How is most of the DNA in an isolated nucleosome in principle available for binding other proteins?
the DNA in an isolated nucleosome unwraps from each end at rate of about 4 times per second, remaining exposed for 10 to 50 milliseconds before the partially unwrapped structure recloses.
What post translational modifications can histones undergo?
Acetylation - of Lysine (K)
Methylation - of Lysine (K) and Arginine (R)
Phosphorylation - of Serine (S), Threonine (T) and Tyrosine (Y)
Citrullination - of Arginine (R)
Ubiquitination - of Lysine (K)
Sumoylation - of Lysine (K)
What processes are modifications important in?
Mitosis
Meiosis
Replication
Repair
Apoptosis
Transcription
Which histones can be acetylated?
Lysines in H2A, H2B, H3 and H4
Which histones can be methylated?
Lysines and arginines in H3 and H4
When does PMT of histones in nucleosomes occur?
in situ (i.e. whilst histones are bound to DNA)
What does PMT of histones in nucleosomes correlate with?
The functional status of the relevant genomic locus
What do PMT of histones define?
An epigentic histone code and corresponding functional landscape
What does the epigenetic histone code define?
the binding potential of chromatin associated factors
Acts as markers for other proteins to come in and bind to tails of histones
Then those proteins determine whether gene is activated or inhibited
What enzymes add acetyl groups to lysines?
Histone acetyltransferases (HATs)
What are examples of HATs?
yeast Gcn5 and its homologues, CBP, PCAF and TAF250
How do HATs often exist?
In large multi-subunit complexes recruited by TF activators eg nuclear receptor co-activator complexes
How do HATs acetylate lysines?
By transferring an acetyl group from acetyl-coenzyme A (acetyl-CoA) to the ε-amino group of the lysine residue
What enzyme removes acetyl groups from lysine?
Histone deacetylases (HDACs)
Examples of HDACs?
yeast Rpd3, human HDAC1
How do HDACs often exist?
Within large multi-subunit complexes
How is acetylation associated with transcriptional activation?
Influences the structure of chromatin and TF access
Removes the positive charge of lysine
Less cross-binding of histones
Relaxed stated
What kind of activity does CLOCK contain?
HAT - can acetylate H3 and H4
What does CLOCK do?
Acetylates H3 and H4
This opens the core tetramer making the DNA accessible for transcription
Hence, CLOCK relaxes the chromatin in addition to recruiting TFII+pol-II
CLOCK keeps the basal promoter region open for TFII+Pol-II accessibility
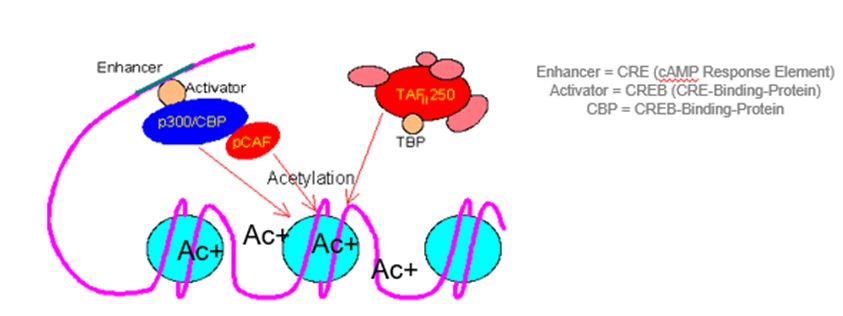
How is CREB and CRE another example of acetylation?
CREB (CRE-binding protein) is an activator and binds to the enhancer CRE (cAMP response element)
CREB then binds to p300 and CBP (CREB binding protein)
p300/CBP can interact with a variety of transcriptional regulators such as pCAF (a histone acetyltransferase) and TBP (which recognizes the promoter [TATAA])
TBP is associated with TAFII250 which can also acetylate DNA
During transcription, they are all assembled at the promoter region.
Histone acetylation by p300/CBP/pCAF and TAFII250 facilitates assembly of the pre-initiation complex to drive transcription
What enzymes methylate lysines?
Lysine methyltransferases (KMTs)
What methylation state is involved for activation?
Hypomethylation (monomethyl-lsyine)
What methylation state is associated with transcriptional repression?
Hypermethylation (dimethyl or trimethyl)
Used as a tag for other proteins to come and block
What is an example of human KMTs?
SUV39H1 & SUV39H2 are human lysine methyltransferases that specifically methylate histone H3 on lysine 9 (H3K9) to generate H3K9me2/3
Associated with repression!
How do KMTs methylate lysines?
KMTs use S-adenosyl-L-methionine (SAM) as a donor to transfer a methyl group to the epsilon-amino group of a lysine residue
Methylation can add 1, 2 or 3 methyl groups to the NH3+ residue of lysine
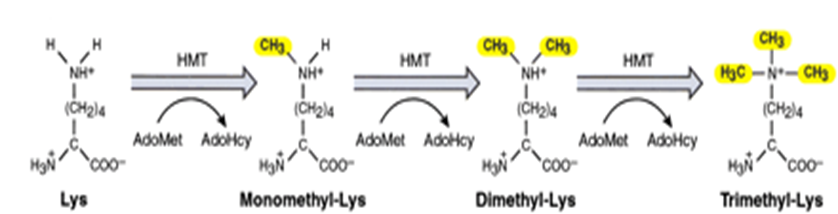
It does this by replacing a proton (H+) with CH3 each time
What enzymes demethylate lysines?
Lysine demethylases (KDMs)
What enzymes methylated arginines?
Protein arginine methyltransferases (PRMTs)
How is monomethyl-arg formed?
By PRMT type 1 or 2 catalyze the transfer of methyl groups from S-adenosylmethionine (AdoMet/SAM) to nitrogen atom of arginine residues in histones
How is asymmetric dimethyl-arg formed?
Methylation of monmethyl-arg by type I PRMT
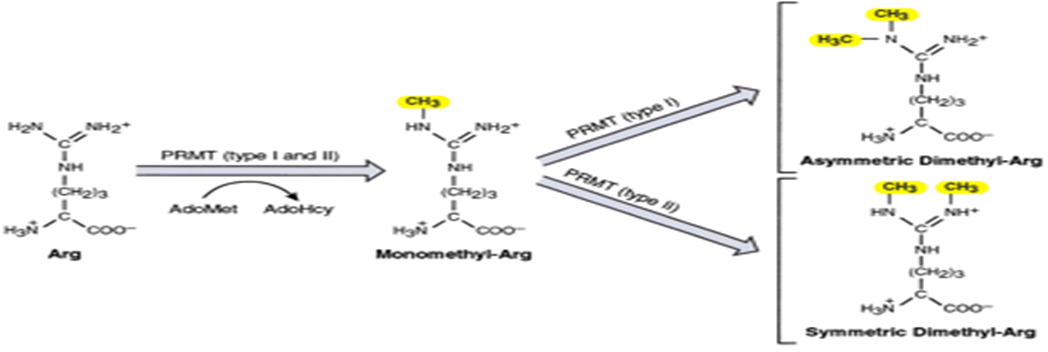
How is symmetric dimethyl-arg formed?
Methylation of monomethyl-arg by type II PRMT
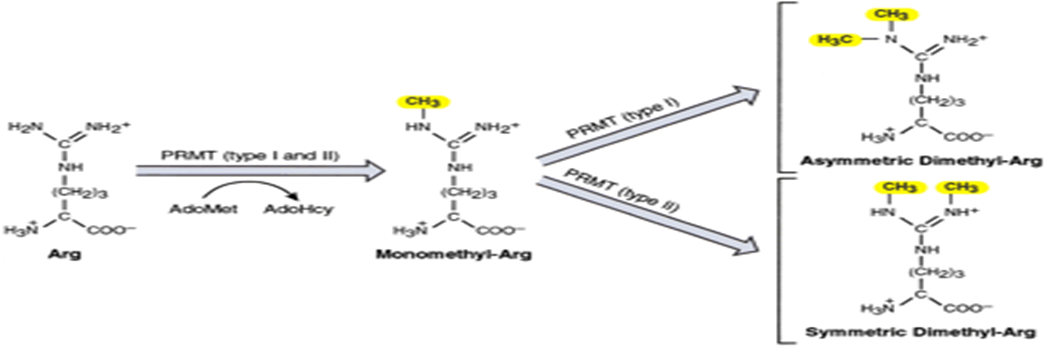
What does PRMT1 do?
Methylated H4R3 for transcriptional activiation
What does PRMT4 do?
Methylted H3R17 and the non-histone protein p300/CBP to trigger its HAT activity for transcription
What is the only Protein Arginine Demethylase in humans?
JMJD6 which demethylates H3R2 and H4R3 for transcriptional repression
What is arginine methylation usally associated with?
Transcriptional activation
Why are specific histone marks found at predictable locations along genes?
Because of different functions they perform
What is in the promoter when the gene is active?
H3K9 is acetylated
What is in the promoter when the gene is inactive?
H3K9 in hypermethylated
What happens in the coding region when the gene is active?
H3K9 is hypotheylated (mono-methyl)
What is the difference between active and inert genes?
They have very different patterns of histone PT modification
What do modified histone tails do?
alter chromatin function by driving association with specific bound regulatory proteins
What do H3K4me3 and H3K9me3 do?
bind to and repress PHD finger domain proteins – eg. ING2, BPTF
What do H3K9me2/3 and H3K27me2/3 do?
bind to and repress chromodomains – eg. HP1
What do general-Kac and H4K16ac do?
Bind to and activate bromodomains eg PCAF
What do general-Kac and H4K5acK12ac do?
Bind to and activate double bromodomains eg hTAF1
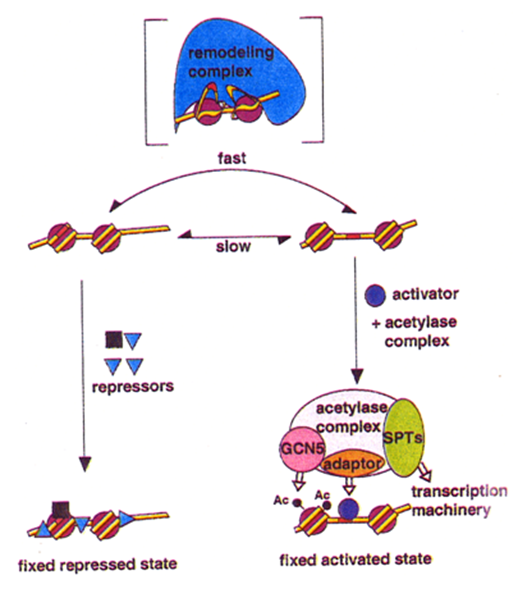
What are chromatin remodelling complexes?
Control transitions between chromatin states
eg o Want enhancers to be in linker regions so it is more accessible for factors to bind
What do chromatin remodelling complexes bind to?
Both the protein core of nucleosome and the ds DNA around it
What are the 2 major classes or chromatin remodelling complexes?
ATP-dependent chromatin remodelling histone sliding
Histone acetylase/deacetylase containing complexes
What is an example of ATP-dependent chromatin remodelling?
SWI/SNF and NURF
What is an example of Histone acetylase/deacetylase containing complexes?
SAGA and PCAF
What is euchromatin?
Active gene
How do ATP dependent chromatin remodelling complexes and histone acetylase complexes work?
By using the energy of ATP hydrolysis to move DNA relative to the core, this subunit changes the structure of a nucleosome temporarily, making the DNA less tightly bound to the histone core.
Through repeated cycles of ATP hydrolysis, the remodeling complexes can catalyze nucleosome sliding , and by pulling the nucleosome core along the DNA double helix in this way, they make the nucleosomal DNA available to other proteins in the cell
Remodelling allows histone sliding - reveals binding sites
Transcription factors can then recruit histone acetylases to keep the chromatin open (i.e. relaxed and accessible)
Can also hide binding sites to repress
Dynamics of this process is central to gene expression
What is FACT?
facilitator of active chromatin transcription
What does FACT do?
It is a heterdimeric factor which removes the H2A-H2B dimers ahead of the polymerase so that the histones disassemble to allow the Pol-II to advance and transcribe the DNA
What happens once Pol-II has passed?
FACT reassemble the histones to reform a new nucleosome that is situated further upstream (i.e. 5’) from its start position
What are tranacription activation domains?
Contact other proteins which facilitate recruitment of a functional transcription pre-initiation complex
They are on enhancer elements and contact general TFs and trigger RNA pol to start transcribing
What are the common types of activation domains?
Proline rich
Glutamine-[Q]-rich
Acidic = glutamate-[E] and aspartate-[D]-rich
How do activators work?
Activators work at a distance by bringing proteins associated with key regulatory elements into a single protein super-complex at promoters
This results in DNA looping and drives the efficiency of pre-initiation complex formation
What are LCRs?
Locus Control Regions (LCRs) act like enhancers but work over longer distances.
They contain multiple-binding-sites to allow complexes of TFs to activate multiple linked-genes
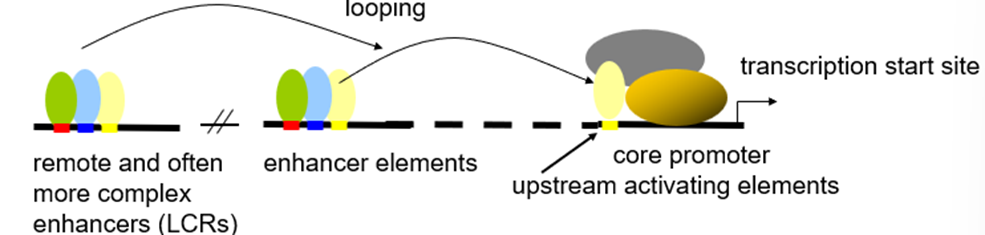
What are insulators?
Provide boundaries for domains within euchromatin.
Chromatin between two insulators is prevented from interacting with intra-chromosomal chromatin outside
What are barriers?
Barriers function like insulators but are more important at the boundary of heterochromatin and euchromatin.
They prevent heterochromatin spreading into regions containing genes
What does the histone code define?
The behaviour of local chromatin domains and correlates with chromatin dynamics and folding into euchromatin and heterochromatin states.
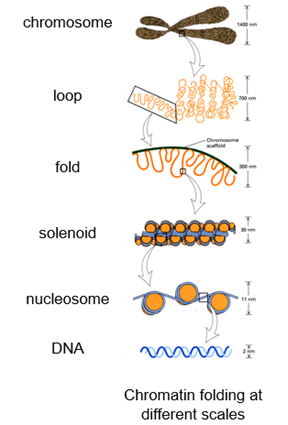
What is the higher folding of DNA?
In the mammalian nucleus chromatin folds and loops to form discrete chromosome territories
CTs are polarised so that euchromatin and heterochromatin occupy discrete zones
Stretches of the genome can be gene-rich or gene-poor
Different chromatin domains fold into distinct ~1 Mbp DNA foci
Regions of DNA that are gene rich end up in a particular domain
Active and inactive higher-order domains are also separated
What are the different chromatin folding scales?
What is the function of nuclear compartments?
Molecular and cell biology approaches show that sites of mRNA synthesis are clustered within factories that typically contain many genes with 5-10 active RNA pol-II complexes
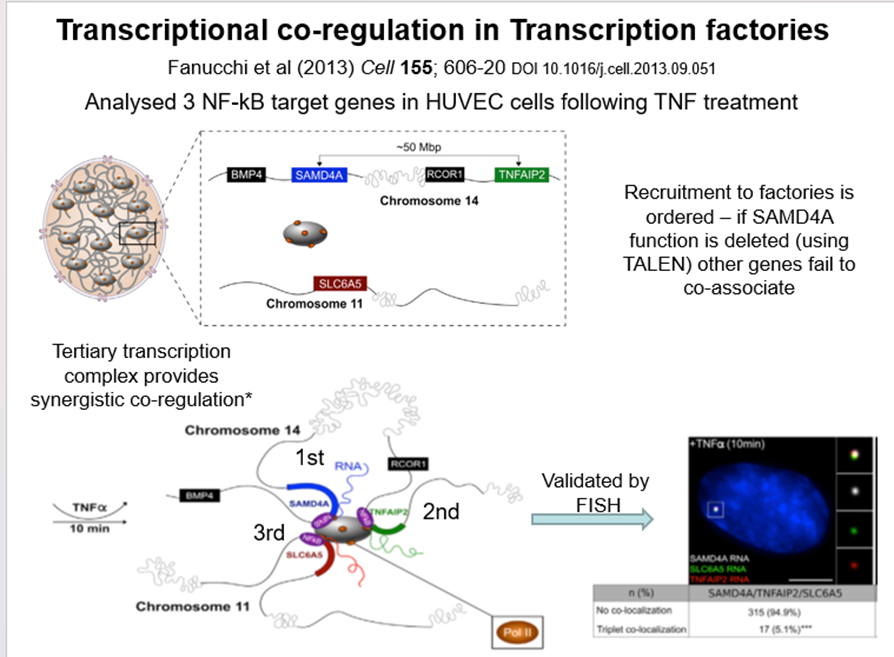
How can transcriptional co-regulation in transcription factories be shown?
3 NF-kappaB target genes analysed in HUVEC following TNF treatment
Recruitment to factories is ordered - if SAMD4A function is deleted (using TALEN) other genes fail to co-associate
Tertiary transcription complex provide synergistic co-regulation