Chromatin
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
Epigenetics diagram
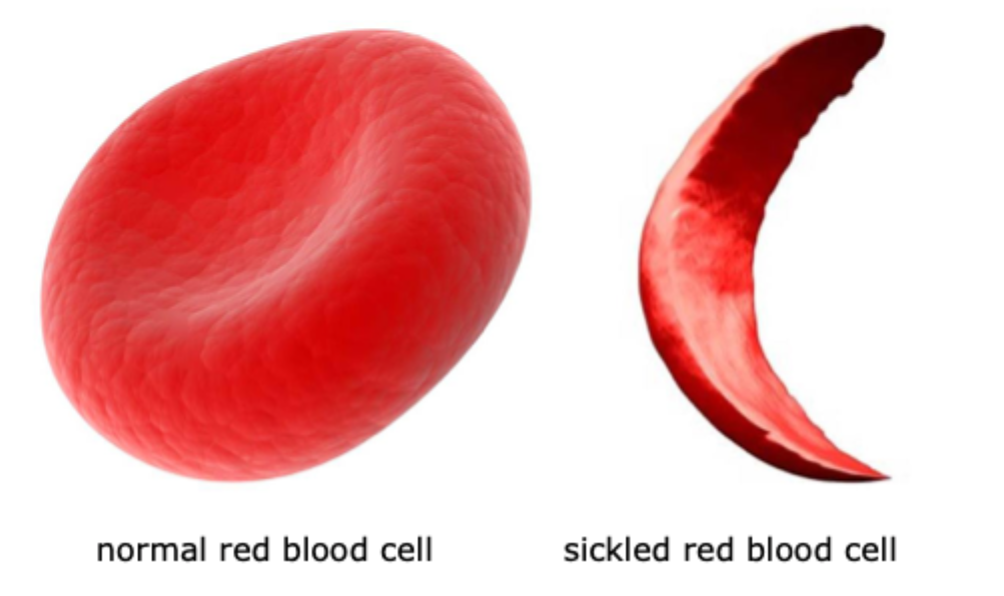
Haemoglobinopathies- normal RBC vs tickled RBC
Haemoglobinopathies
Most common genetic disorder world- wide
~350,000 Beverly affected children born each year
e.g. sickle cell disease
a/ b thalassemias
abnormal structure, or reduced expression of either a- and b- like globin chains
Haemoglobin structure
a tetrameter protein consisting of 2 a- and 2 b-like global chains
?
Transcription from a eukaryotic gene
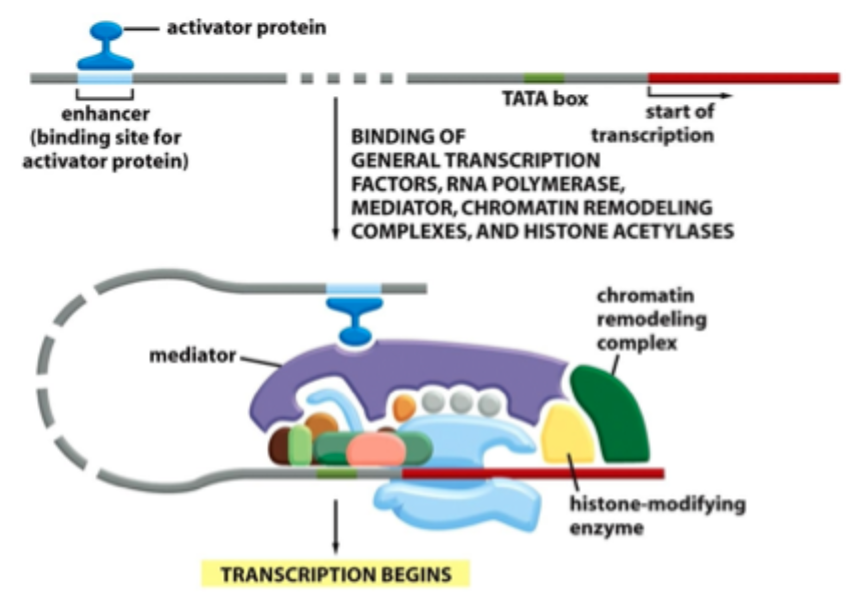
Transcription factors (TFs)- and binding motifs
~650,000 TF binding motifs
~50,000 binding sites for a typical TF
TF motifs are insufficient to predict binding
Accessibility of binding sites changes during development
ES cells, endoderm cells- only 16% overlap of binding sites
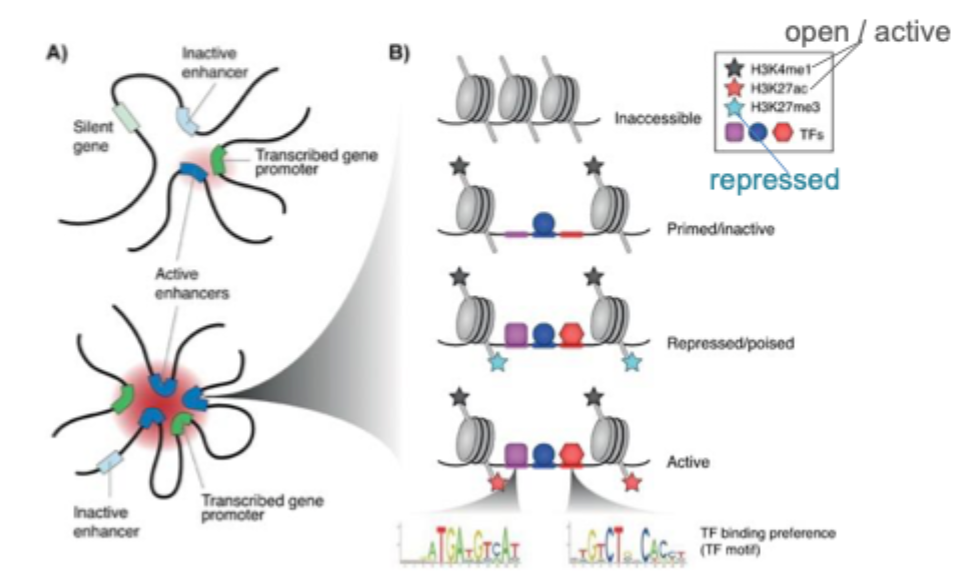
Enhancers
are regulatory DNA sequences
influence (enhance or inhibit) transcription of associated genes
often located remotely from gene promoters
enable correct spatio- t'emportas gene activation
determine cell- type specific expression
can function regardless of orientation, position, and distance
contain TF- binding sites
have both enhancer and promoter activity
generally lower CpG content than ‘classic’ promoters
local chromatin signatures, including H3K4me1 and H3K27ac
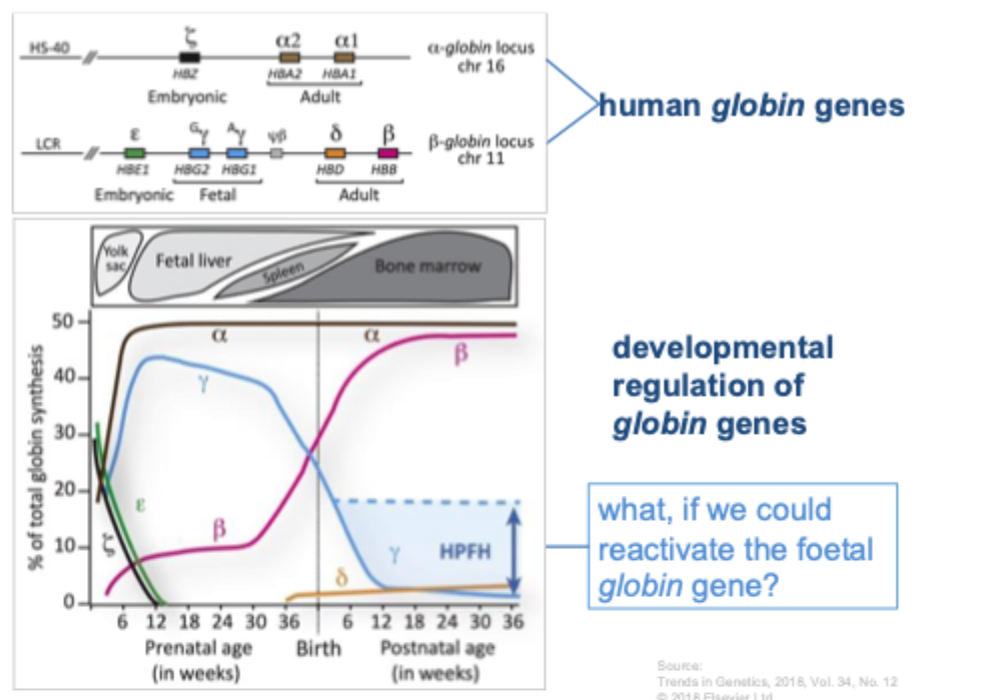
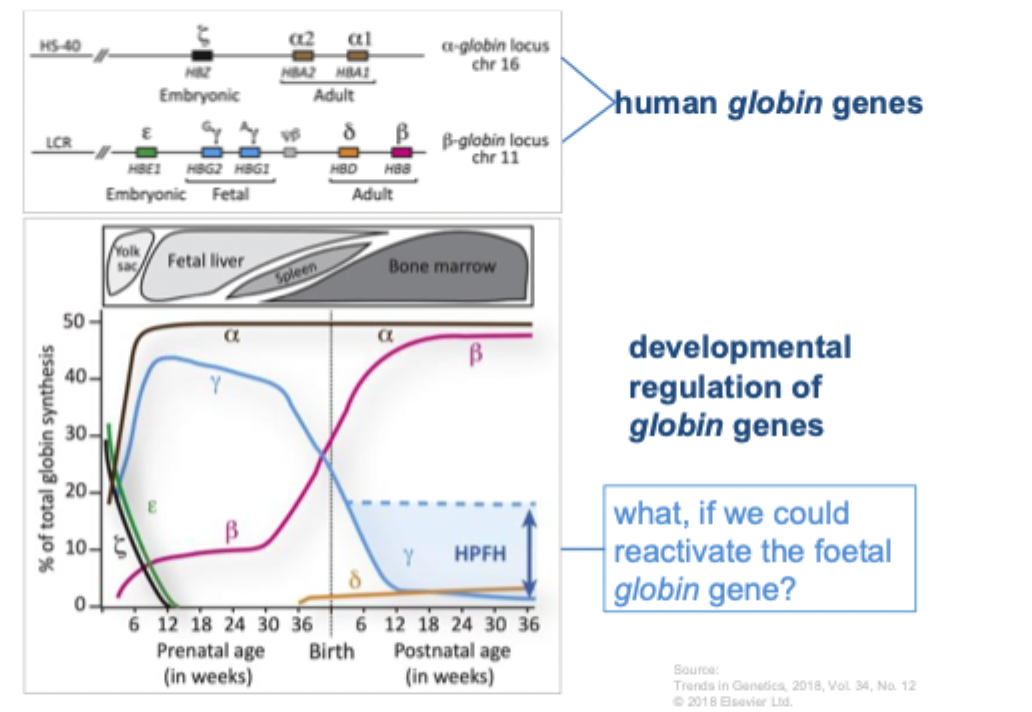
B-globin locus: enhancers, genes, interacting factors
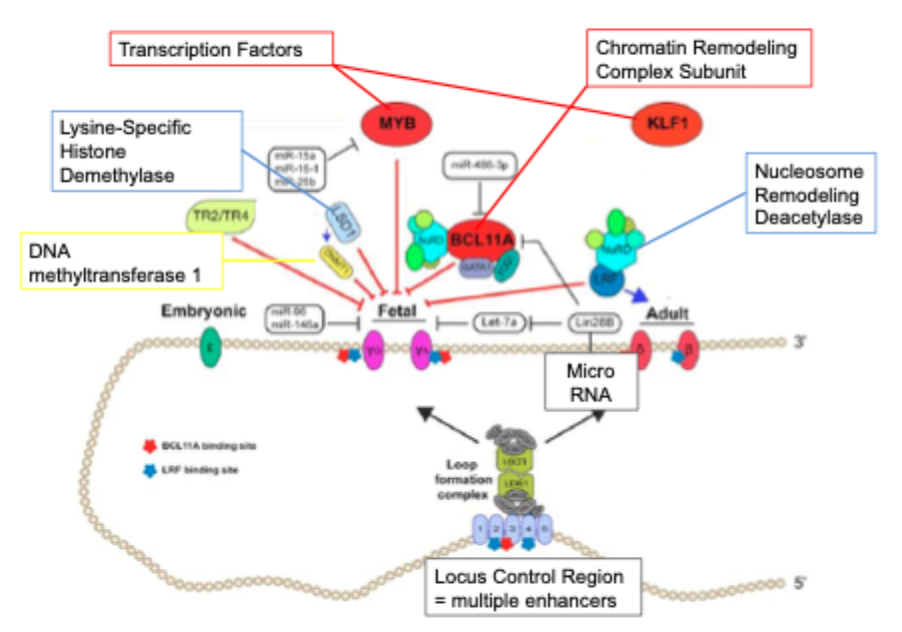
Enhancer- mediated regulation of genes
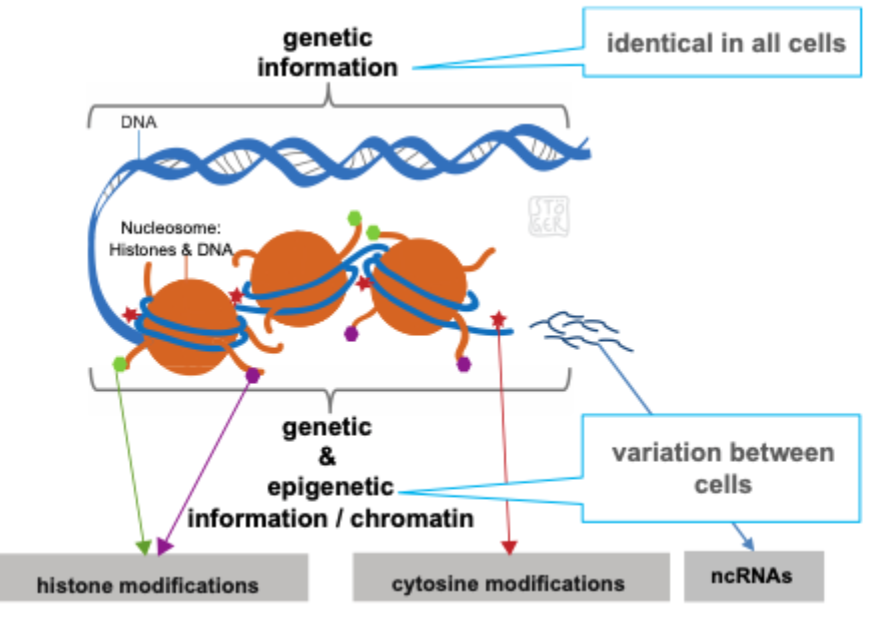
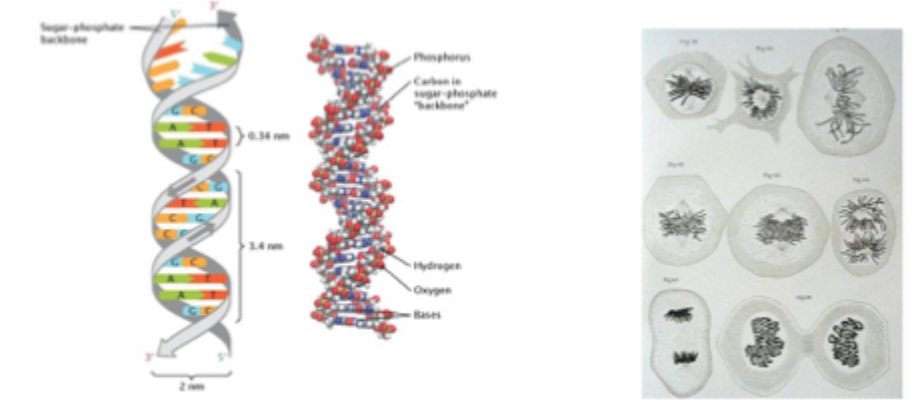
Difference between DNA and chromatin
DNA is not ‘naked’ within cells
DNA is wrapped around proteins = chromatin
Limited accessibility for transcription factors
Chromatin
Complex of DNA, histones and non- histone proteins found in the nucleus of a eukaryotic cell- the material of which chromosomes are made
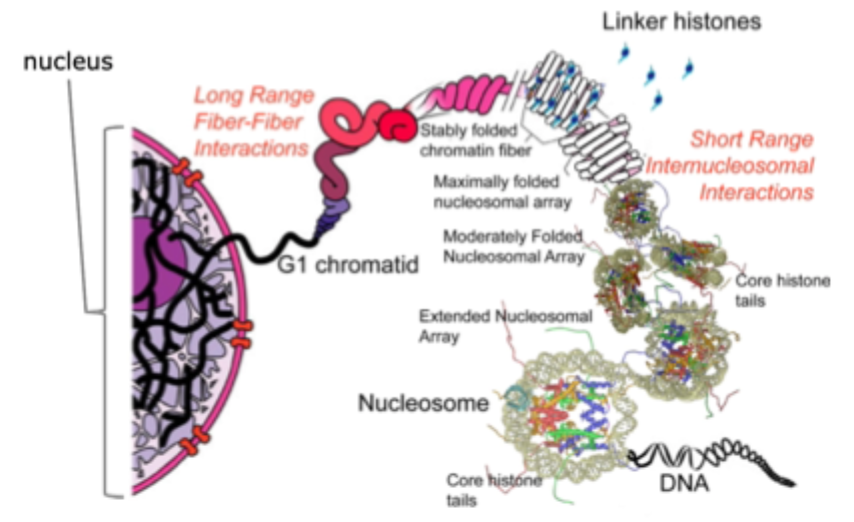
Chromatin
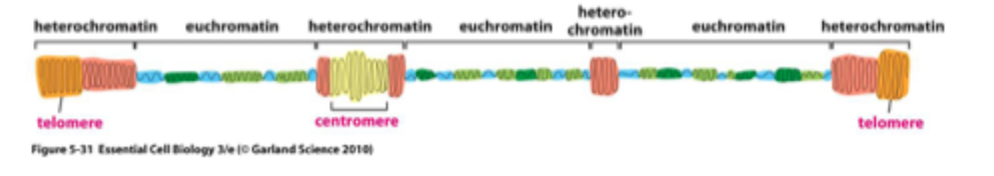
Chromosomes are organised into open regions with active gene transcription (euchromatin) and closed regions where genes are repressed (heterochromatin)
Chromatin is an active structure and is remodelled by chromatin remodelling complexes
These can alter the structure of the DNA- histone complex of the nucleosome
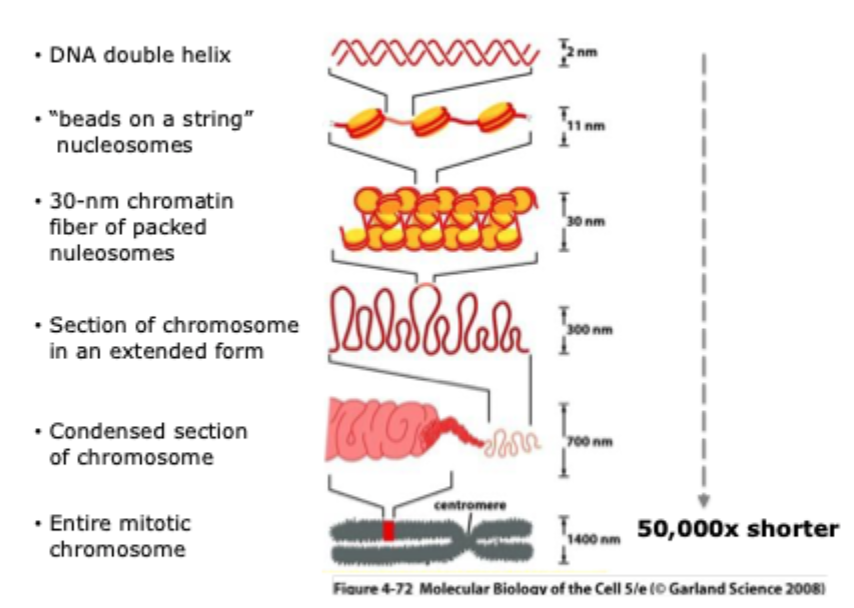
Levels of chromatin packing
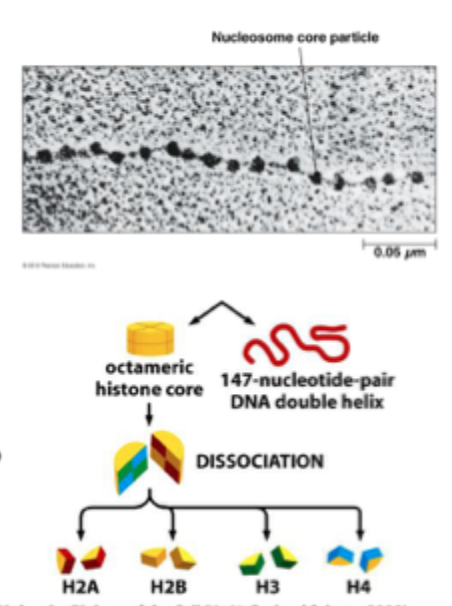
The Nucleosome
Is the fundamental building blocks of chromatin is the nucleosomal core particle
Eukaryotic chromosomes consist of a regularly repeating protein- DNA complex called the nucleosome
147bp of DNA are wrapped (2 turns) around the histone octamer
Each nucleosome consists of a protein octamer, made up of 2 copies each of histones H2A, H2B, H3 and H4
Histone H1: linker histone, binds to the entry/ exit sites of DNA on the surface of the nucleosomal core particle and completes the nucleosome
TAD’s
Segregation of chromatin into active and inactive domains
Units are termed TAD’s
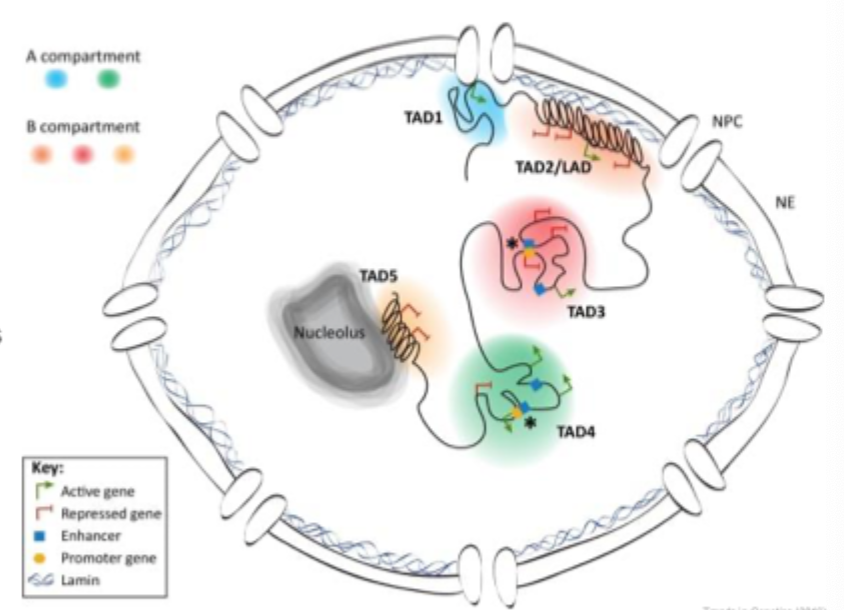
The dynamic nucleus
Within the nucleus DNA is packaged and organised
This is a dynamic and fluid structure which changes with differential requirement of the cell
Discovery of the polycomb- trithorax group (PcG- TrxG) system in drosophila

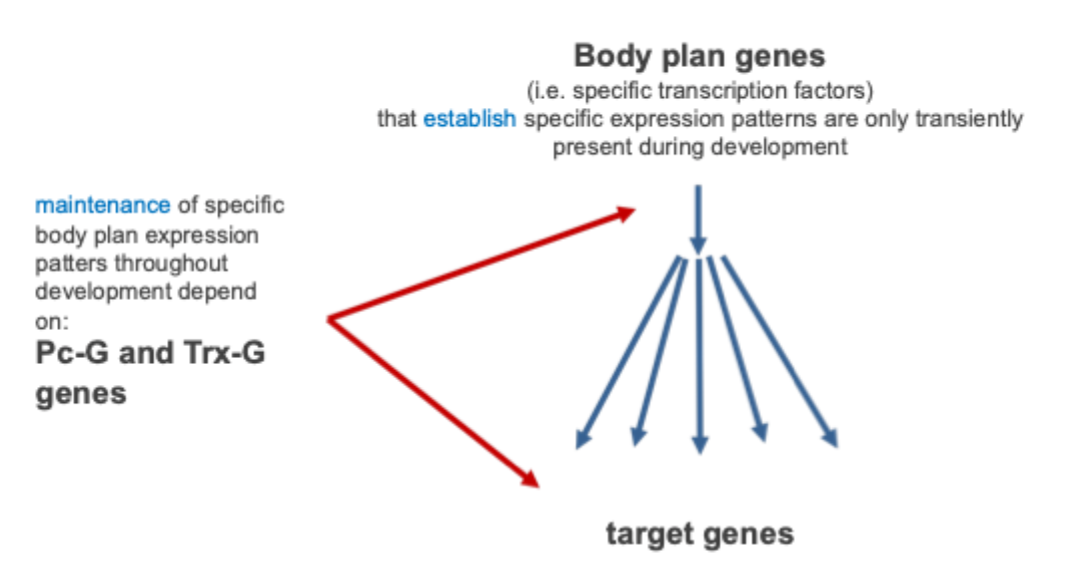
What are body plan genes?
Regulatory genes that control the development of body parts in animals
e.g. expression of pair- rule segmentation gene '“hairy” in drosophila
Spatially restricted DNA- binding transcription factors (i.e. ‘hairy’) control patterned expression of many genes
These direct regulators soon disappear while patterned expression of their targets is maintained
The trithorax group (trxG) are responsible for maintaining expression in the ‘ON’ state and poly-comb group genes (PcG) are responsible for maintaining expression in the ‘OFF’ state
Maintenance of body plan gene expression
TrxG and PcG proteins: concept of cellular memory
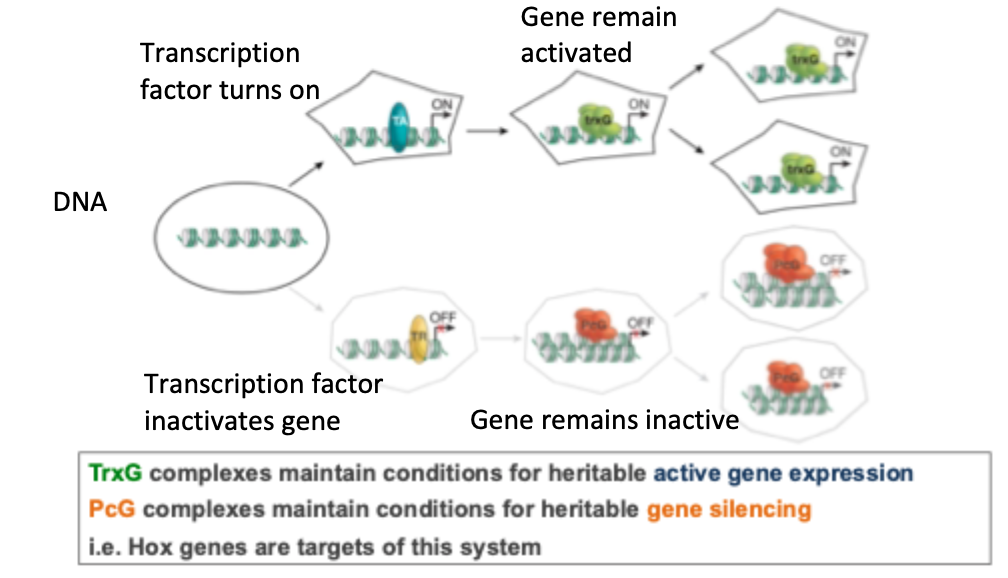
Understood function of Trx/ PcG proteins
Mechanisms of ATP- dependent chromatin remodelling
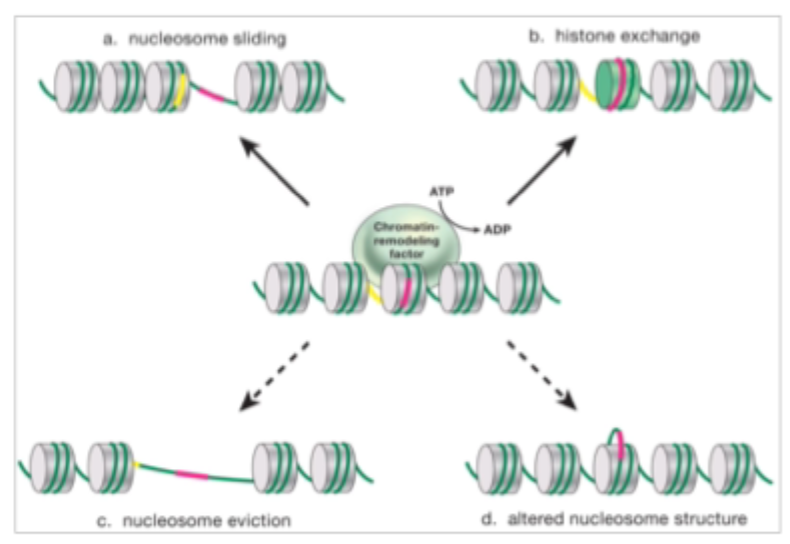
Understood function of Trx/ PcG proteins
Covalent modifications of histones

Histone tails
each of the core histones has an N- terminal tail (amino- terminal) that extrudes from the surface of the nucleosome
Histone tails may help to pack nucleosomes, thereby forming higher order chromatin structures
These histone tails can be subject to several forms of covalent modifications
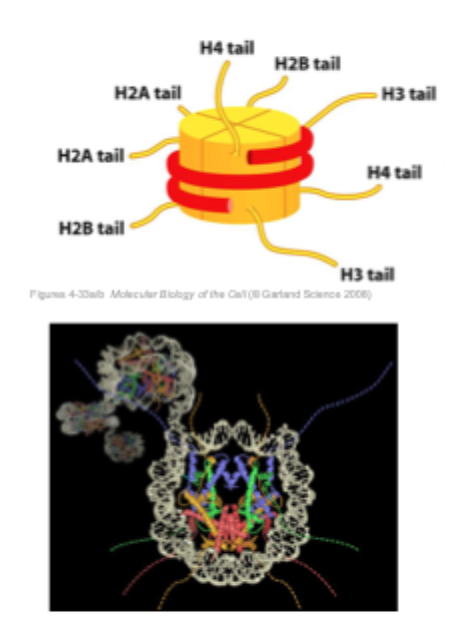
Histone modifications
Acetylation
ADP ribosylation
Biotinylation
Methylation
Phosphorylation
Ubiquitination
Sumoylation
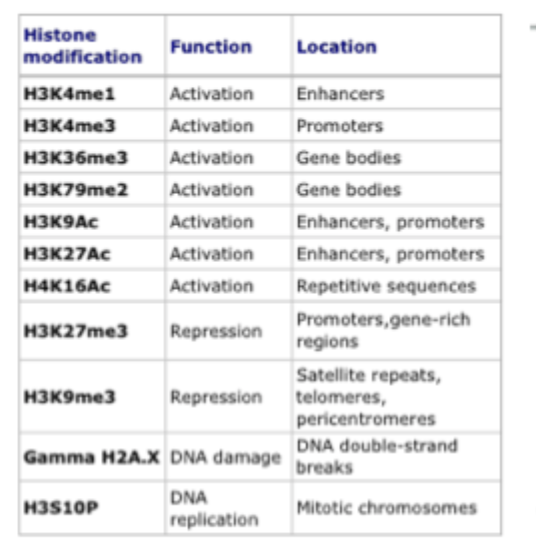
Most common histone modifications and where to find them
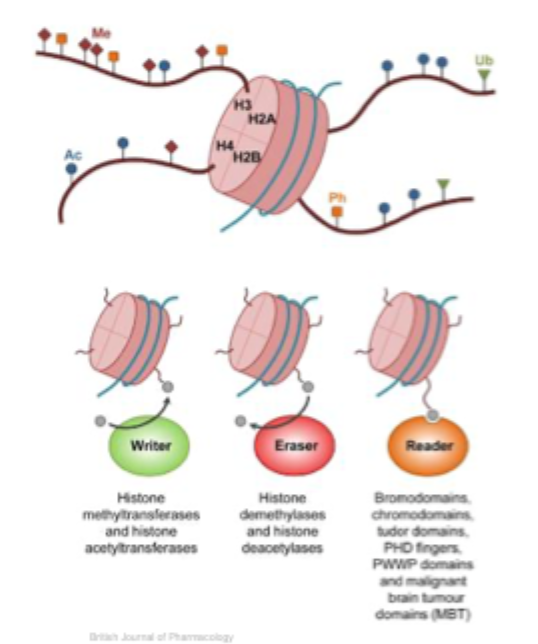
Histone modifications: addition and removal are enzyme mediated
A combination of different modifications can be found on histones- all of these modifications are reversible
Each particular histone modification is created by a specific enzyme
Example: an acetyl group (Ac) is added to specific lysines by a set of different histone acetyl transferases (HAT’s- writers)
The acetyl groups are removed by a set of different histone deacetylase complexes (HDAC’s- erasers)
Sites of histone methylation
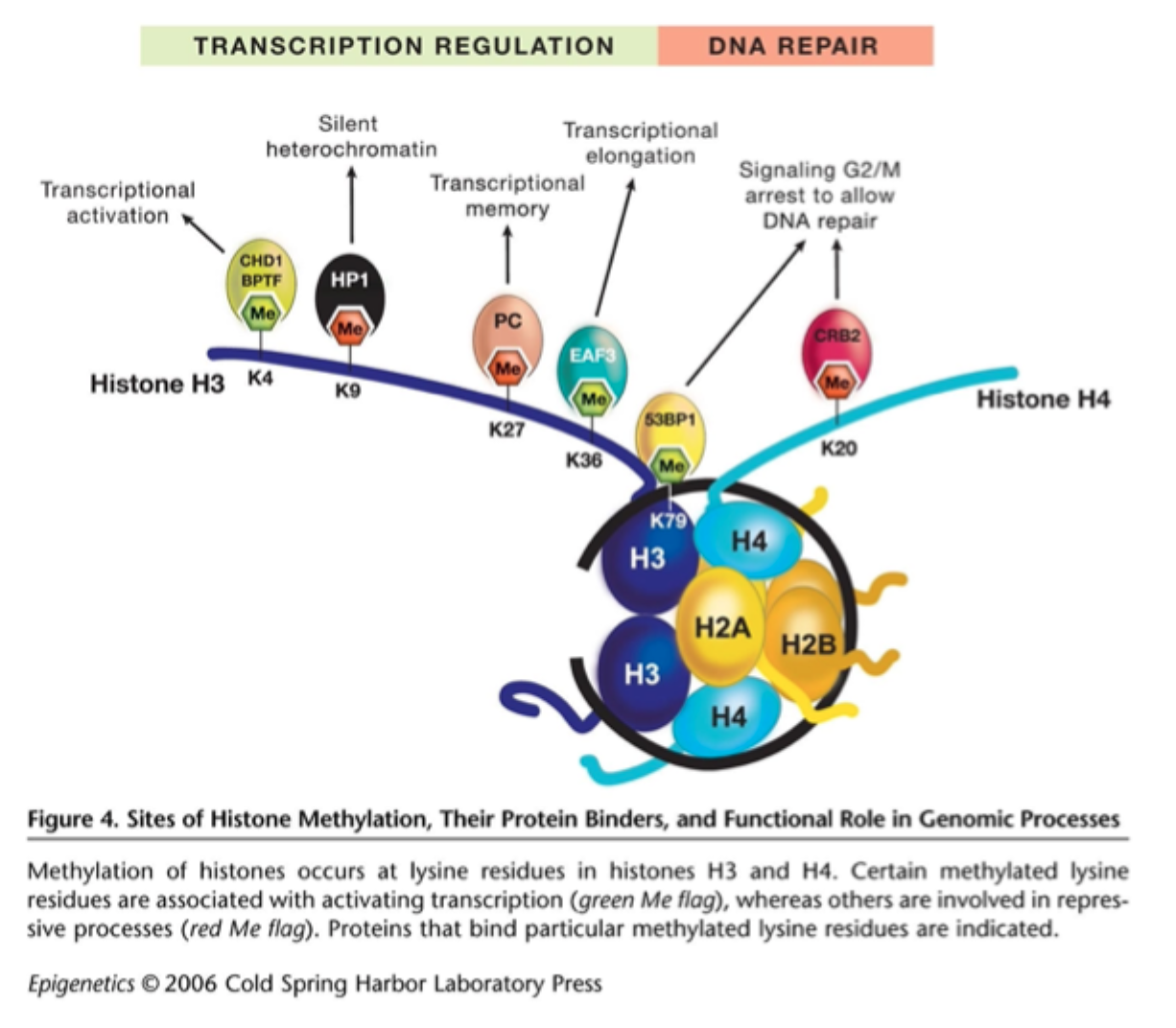
Histone “orthography” writers, erasers and readers
Tri- methylation of lysine residues on histone H3
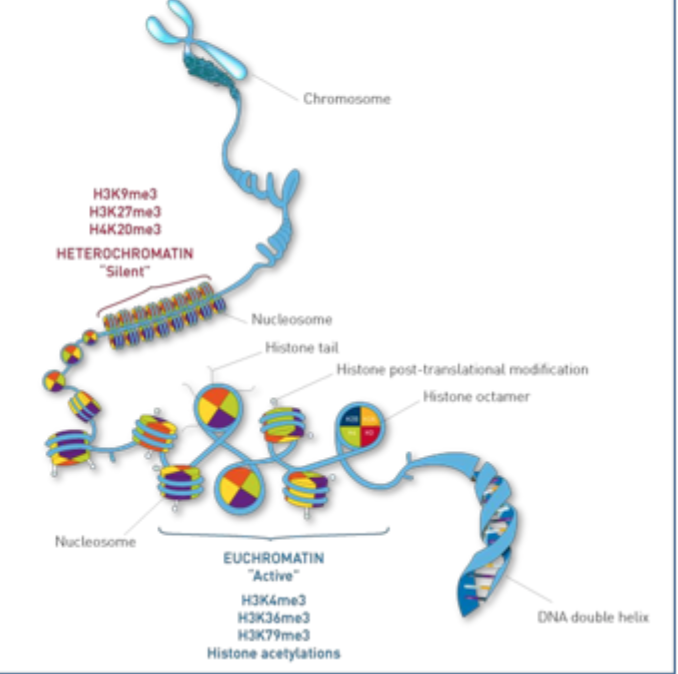
Current concepts
Histone modifications provide functional marks on the nucleosome
Specific marks are recognised (e.g. transcription factors or chromatin model)
There is “cross- talk” between marks (e.g. protein binding to a specific mark can be blocked by modification of an adjacent amino acid)
combinations of marks are important
histone modifications play a role in inheritance of patterns of gene expression
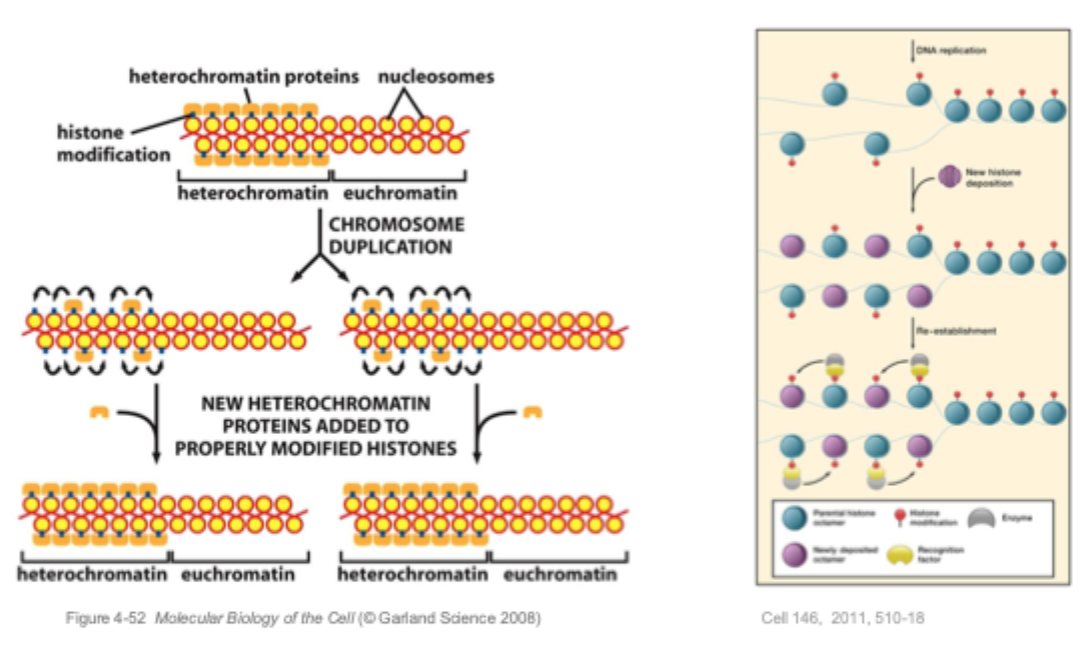
Model: how packaging of DNA can be inherited
During replication, heterochromatin provides a template for its own reassembly.
Not only is DNA replicated, but the chromatin also replicated in a self- templating manner, leading to the propagation of differential chromatin states for multiple generations.
DNA vs Chromatin
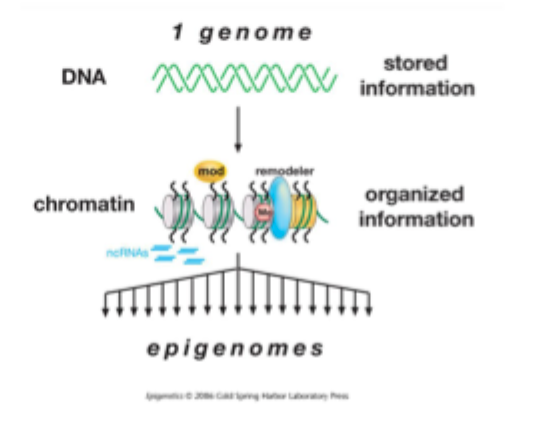
The DNA sequence of an individual is invariant and stores the genetic information
Chromatin, in contrast can vary according to cell type, and in response to internal and external signals it recieves
Diversification of the epigenome occurs during development in multicellular organisms
Epigenome: the overall epigenetic state of a cell
??
Epigenetics
Conrad Waddington coined the term “epigenetic” in the 1940’s
Epi (prefix: on/ besides/ over/ attached to)
with this term he described the branch of biology which studies how genotypes give rise to phenotypes during development
current def: The structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states
Epigenetic landscape
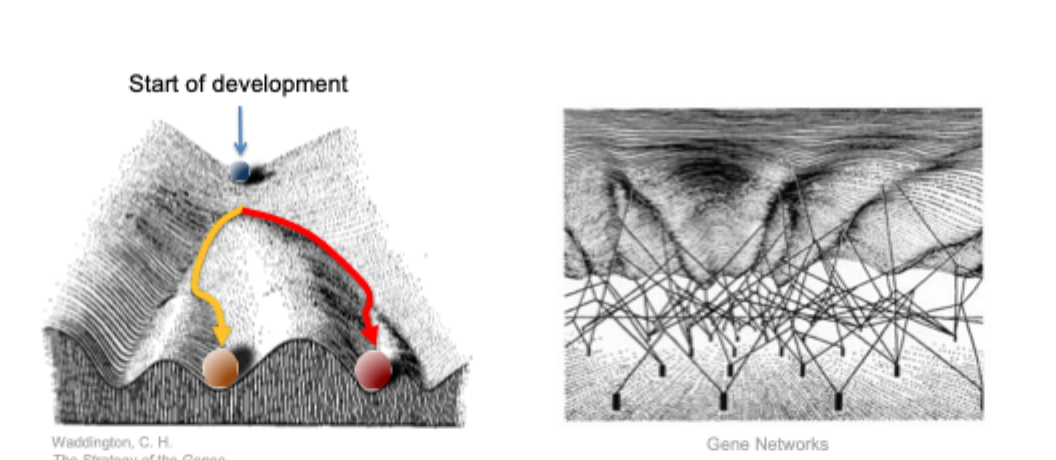
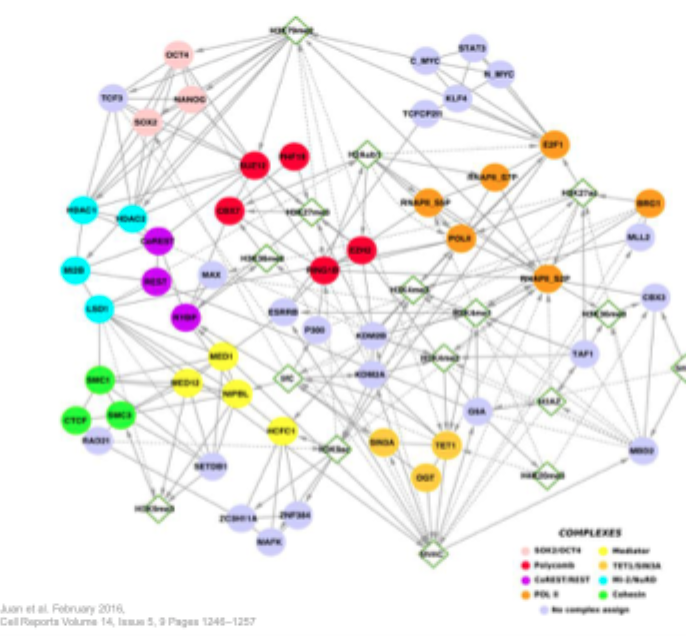
Chromatin communication network in embryonic cells