HMB265
1/438
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
439 Terms
* domestication of plants and animals was a key transition in human civilization
* domestication of dogs from wolves
* domestication of rice, wheat, barley, lentils, and dates from weed-like plants
1. one parent contributes more to an offspring’s inherited traits
2. blended inheritance: traits of parents are blended in their offspring explained single offspring, but not siblings/next gen
* 1st scientist to combine data collection, analysis, and theory to understand heredity
* inferred __**laws of genetics**__ that allowed __**predictions**__ about which traits would appear, disappear, and then reappear
* “experiments in plant hybrids” became __**cornerstone of modern genetics**__
* had the right mind, proper equipment, and appropriate biological material, and INTERDISCIPLINARY EDUCATION
* short generation time
* can be inbred (self-fertilize)
* simple reproductive biology
* small size (easy to grow/breed)
* large numbers o progeny
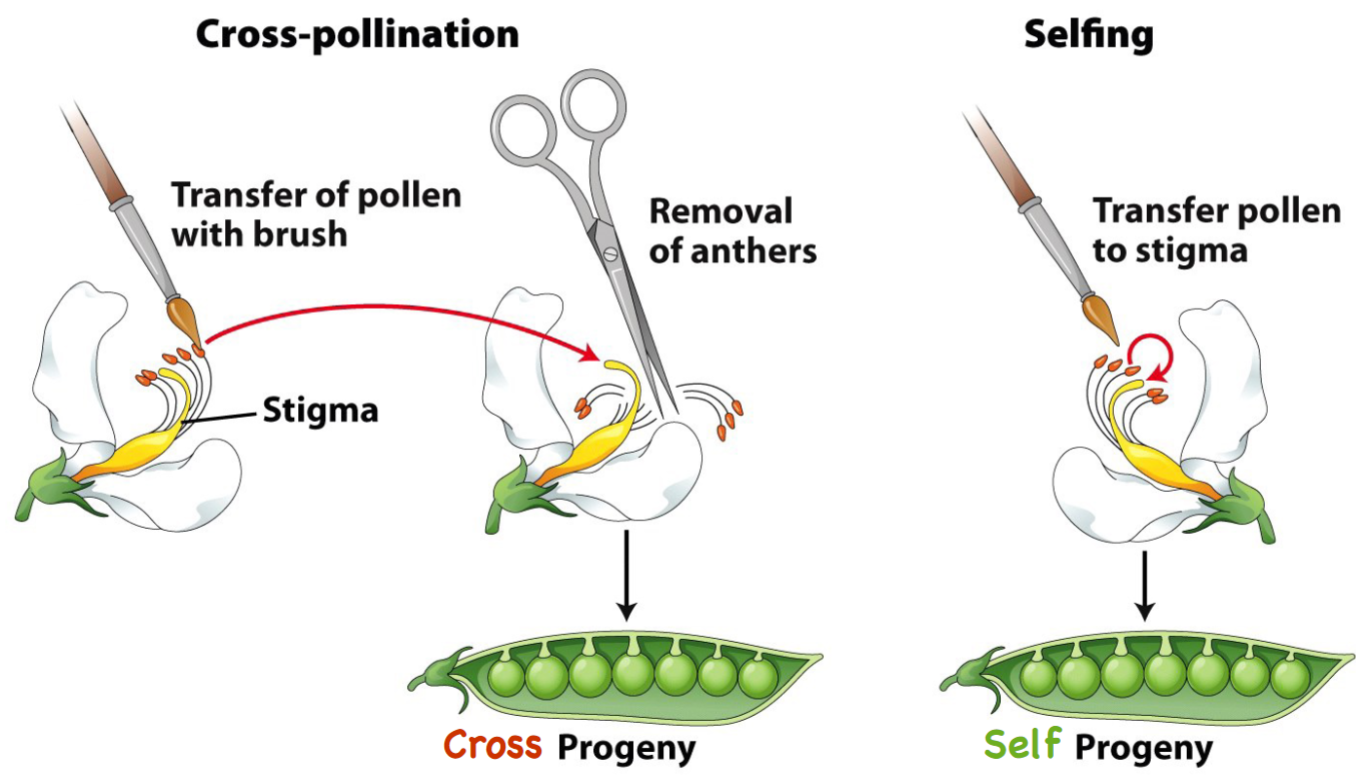
* breeding could be done by __**cross-fertilization/selfing**__ to allow __**inbreeding**__
* could obtain and maintain pure-breeding lines (these always bred true producing the same trait gen upon gen)
* Cross-fertilization enabled reciprocal crosses (to rule out the effect of one parent vs other)
* Large numbers of progeny produced w/in short time (enabling statistical analysis)
* traits remained constant in crosses w/in a line
1. seed colour: green or yellow
2. seed shape: wrinkled or round
3. flower colour: white or purple
4. pod colour: yellow or green
5. pod shape: pinched or round
6. stem length: short or long
7. flower position: at tip of stem or along stem
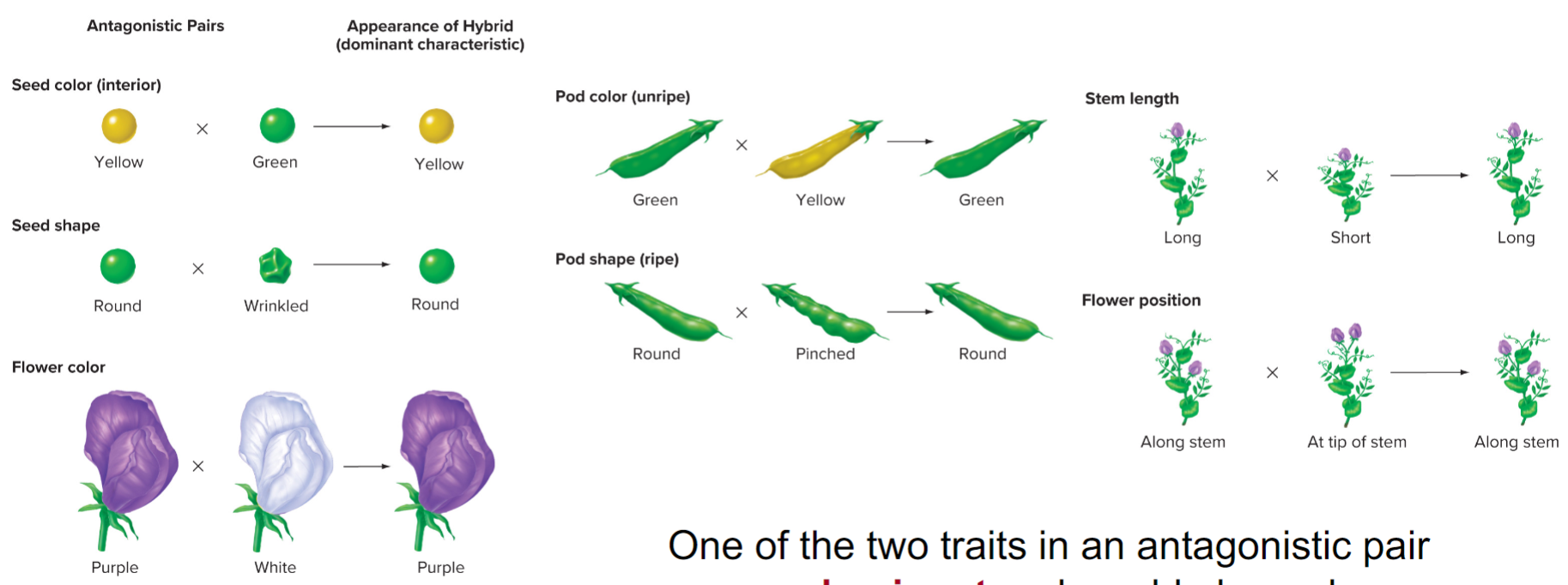
* also revealed that traits could be dominant
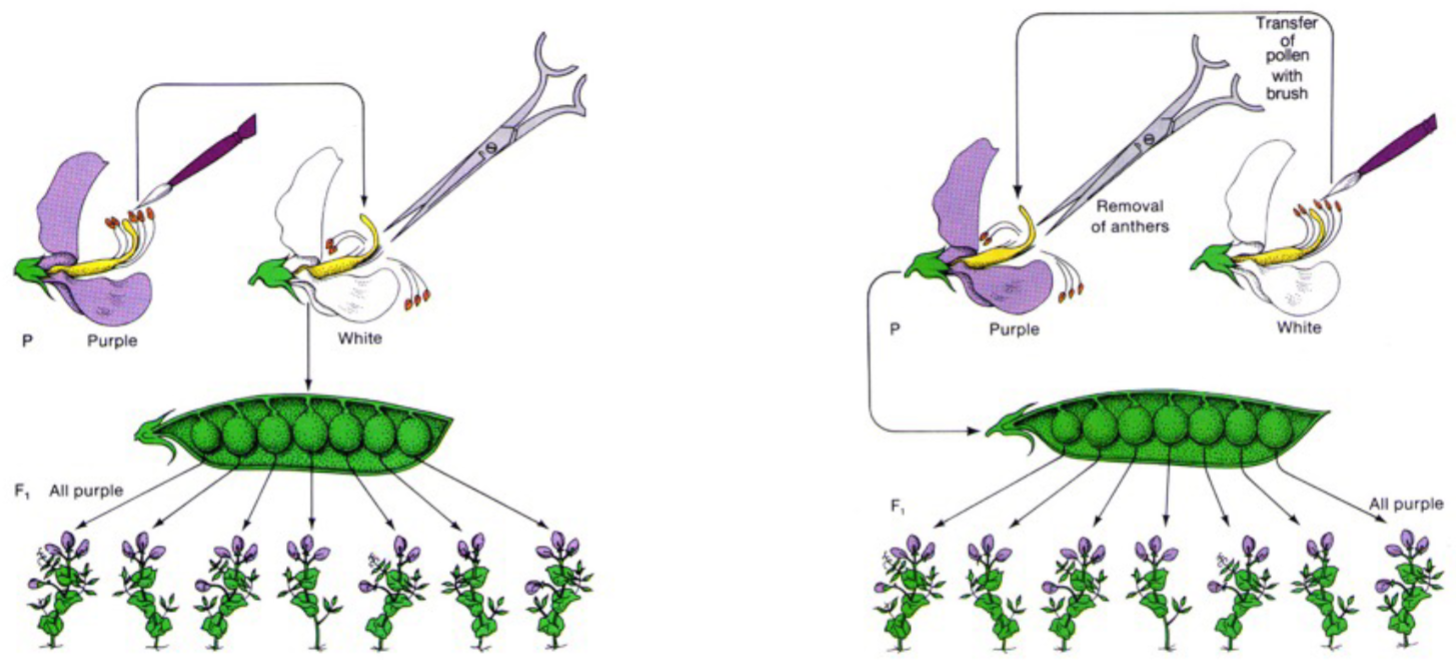
* F1: 1st Filial generation (offspring from parental generation)
* F2: 2nd Filial generation (offspring from F1 gen)
* Self: inbreeding cross that involves individuals that are genetically identical
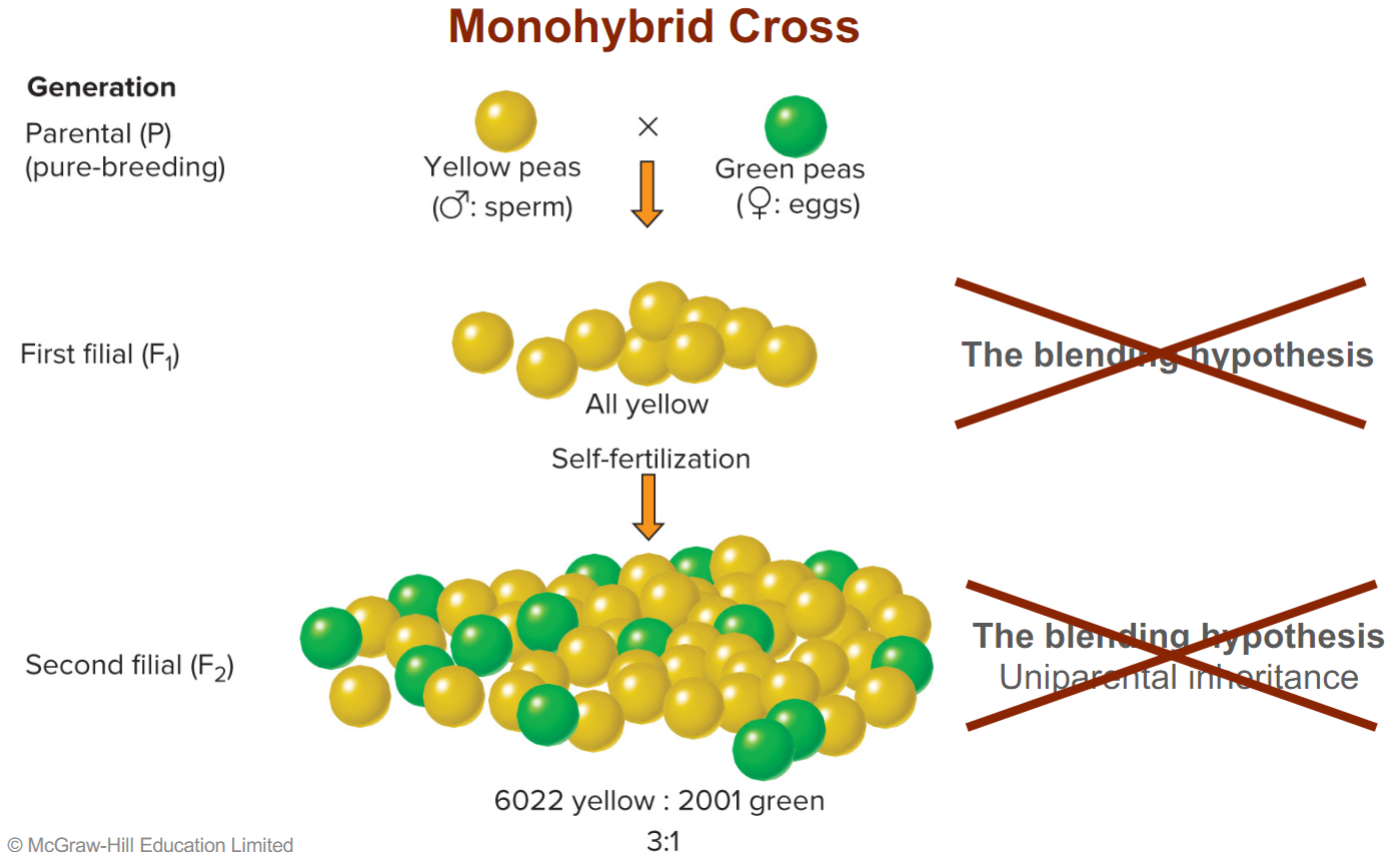
* refuted blending hypothesis in F1 generation, all peas were yellow not yellowish green or greenish yellow
* refuted uniparental hypothesis in F2 gen, because both parents (F1 gen) were yellow peas but F2 gen still had green peas (showing not one parent contributes to offspring)
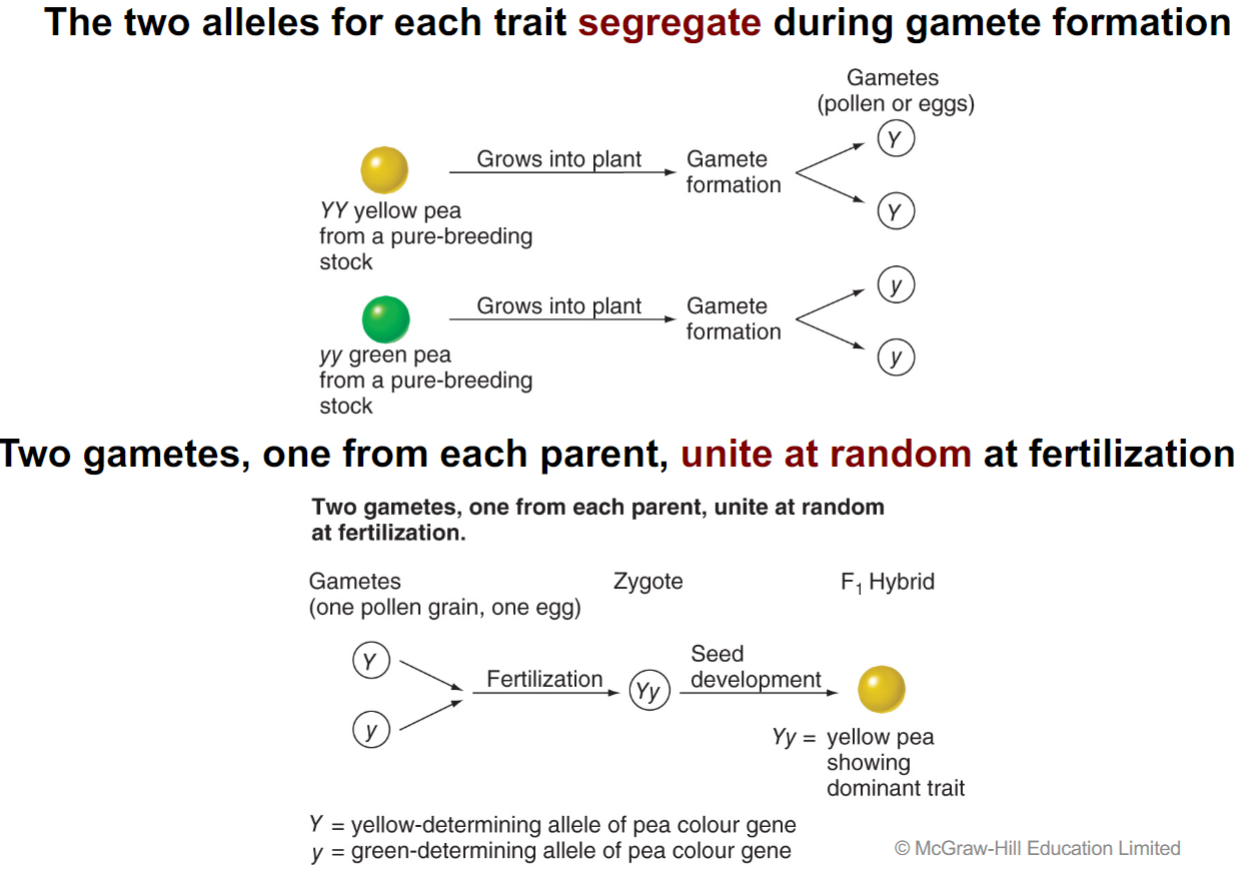
* traits have 2 forms:
1. trait in F1 gen is in __**Dominant**__ form
2. trait that is hidden in the F1 gen is in __**Recessive**__ form
* progeny inherit one unit from each parent
* *now known as* __*“genes*____”__
* Alleles: alternative forms of single locus
* Dominant: allele that manifests itself regardless of other allele that is present ==(indicated using upper case letter)==
* Recessive: allele whose effect is “masked” when dominant allele is present. __**Appears only when other alleles are recessive**__ ==(indicated using lower case letter)==
* Homozygous: when both alleles at given diploid locus are the same (could be dominant/recessive)
* Heterozygous: has 2 diff alleles present at diploid locus (dominant allele w/ - represents unknown genotype, ex: A-)
* Hybrid: derived from 2 diff parents
* monohybrid: 1 hybrid locus
* Dihybrid: 2 hybrid loci
* True-breeding: homozygous at the loci/locus in question
* 2 gametes, one from each parent, unite at random at fertilization
* mendel’s first law incorporates the fact that his results reflected basic rules of probability
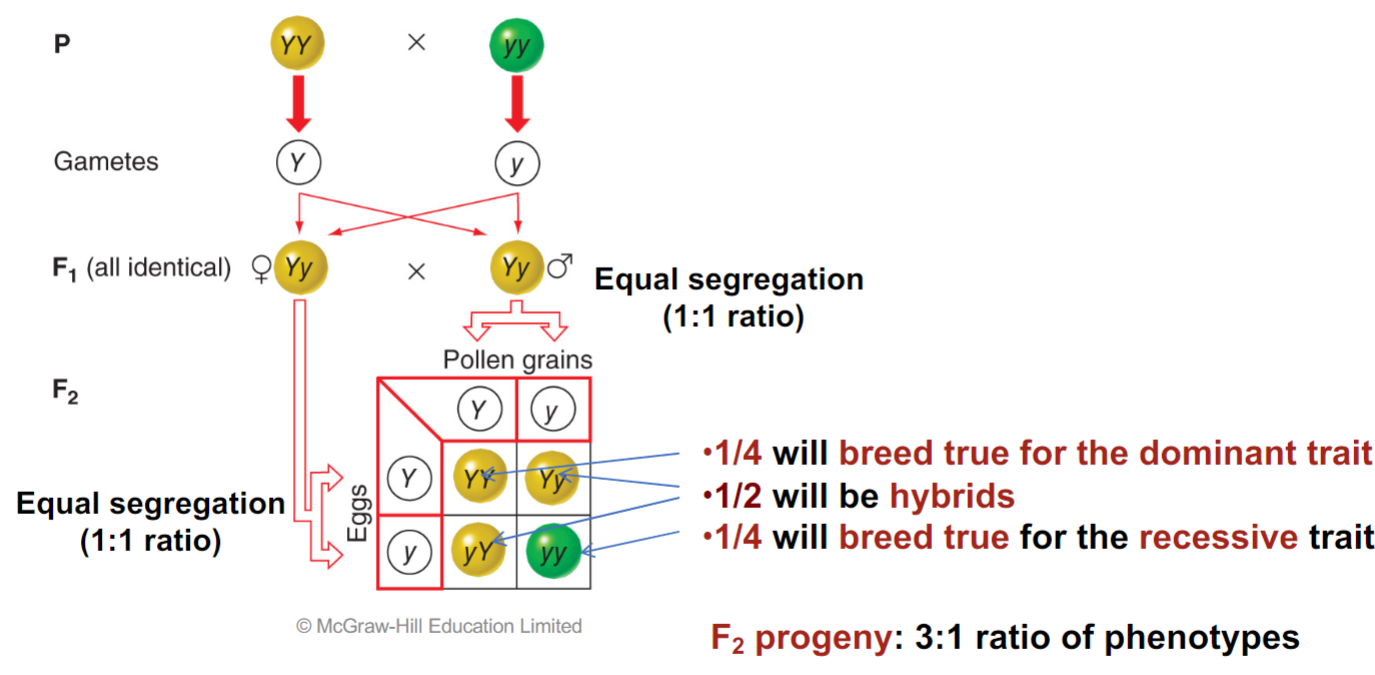
* probability of 2 __**independent events**__ occurring together is the product of their individual probabilities
* Probability of event 1 __**AND**__ event 2 occurring?
* P(1+2) = P1 x P2
Sum rule:
* probability of either of 2 mutually exclusive events occurring is the sum of their individual probabilities
* probability of event 1 __**OR**__ event 2 occurring?
* P(1/2) = P1 + P2
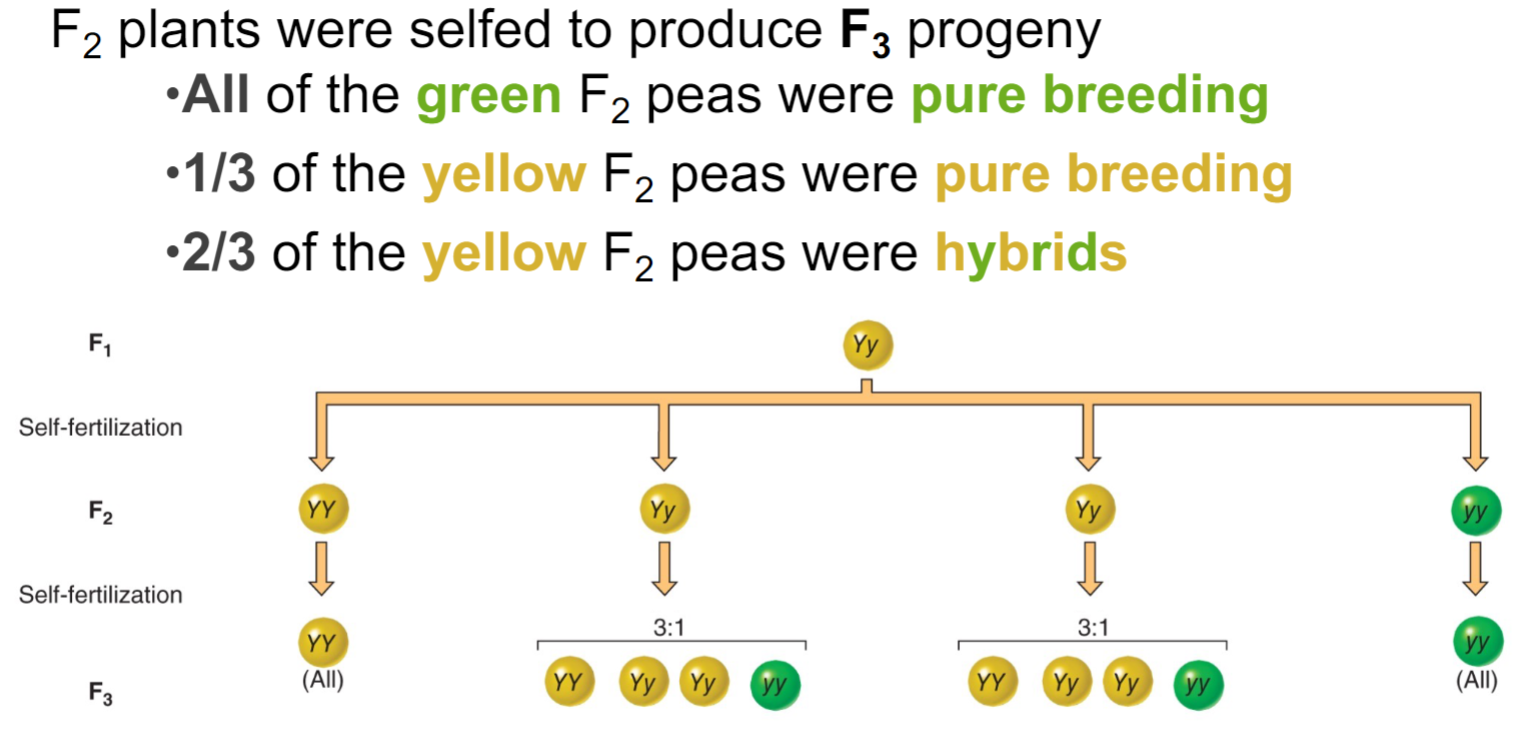
* Phenotype: observable characteristic of an org
* used to discriminate btwn homozygotes dominant and heterozygotes
* testcross = unknown genotype x homozygous recessive
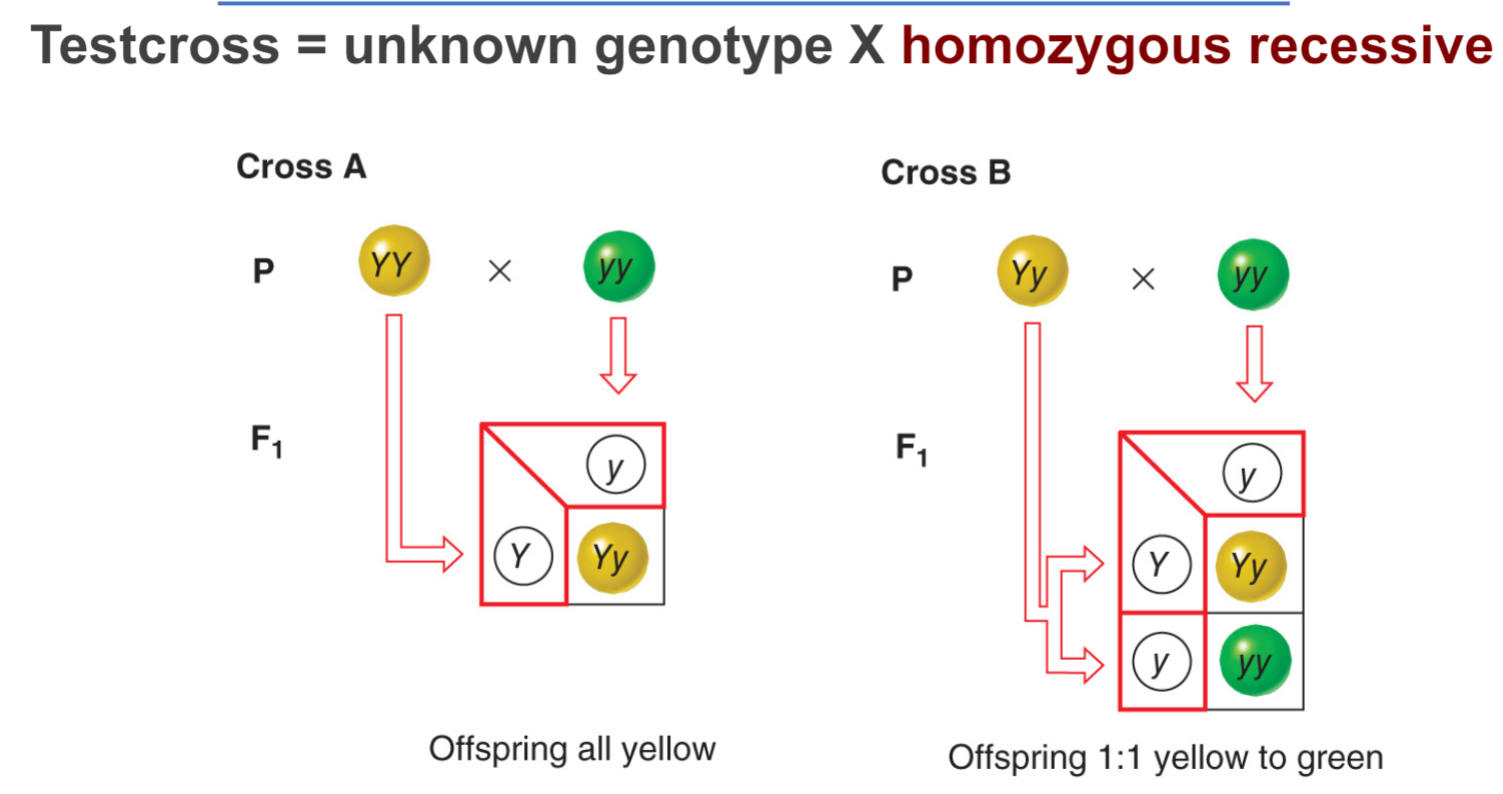
* mendel asked whether all the F2 progeny would be __**parental types**__ (yellow round and green wrinkled) or would be some __**recombinant types**__ (yellow wrinkled and green round)
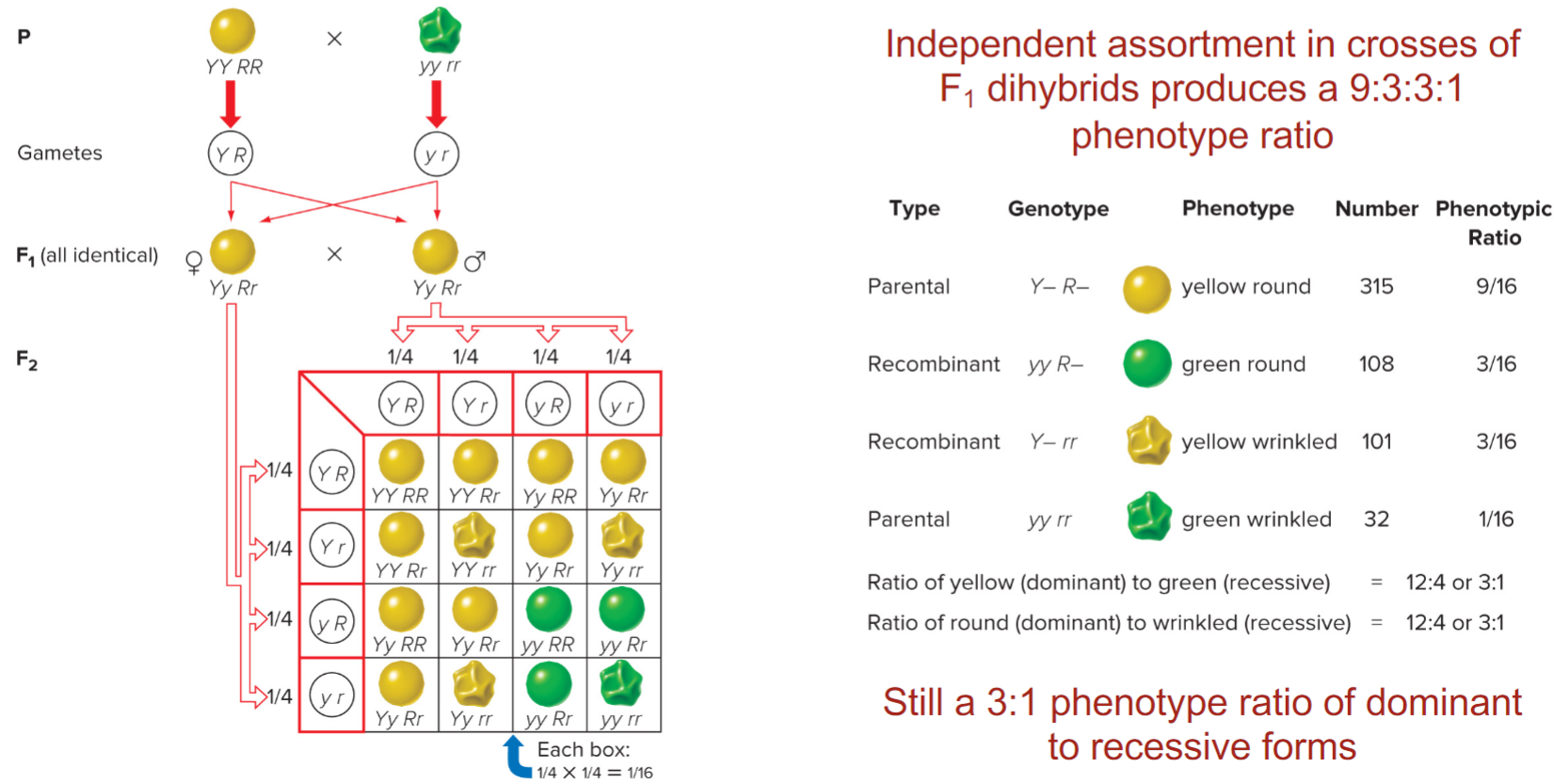
* Y is just as likely to assort w/ R as it is w/ r
* y is just as likely to assort w/ R as it is w/ r
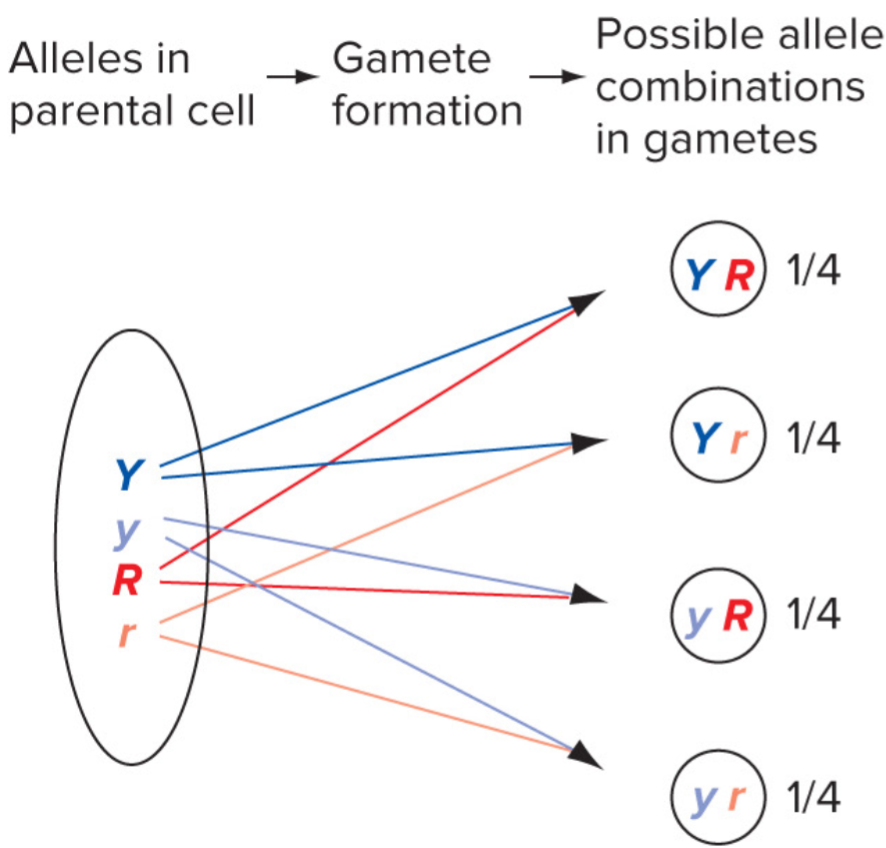
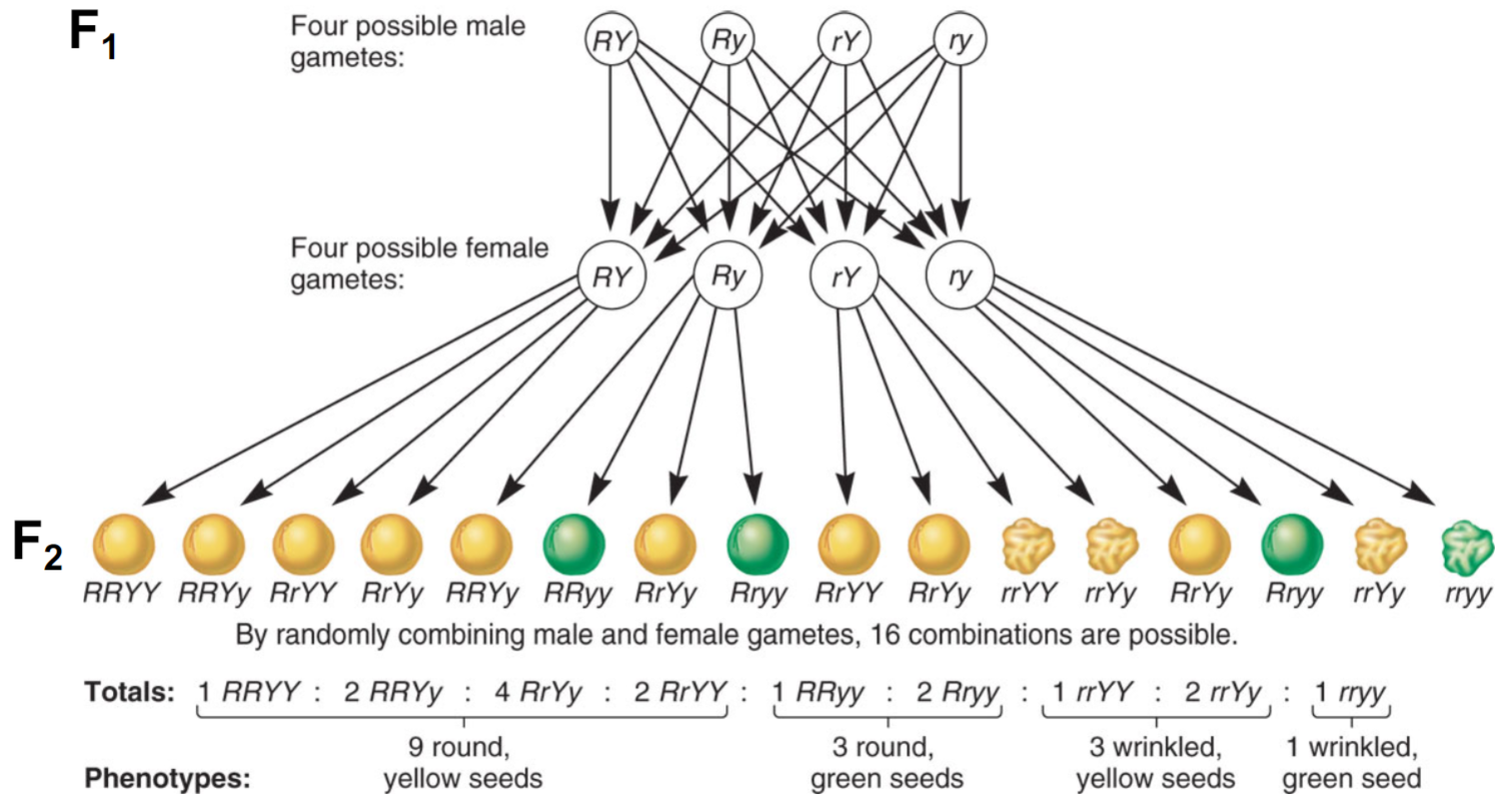
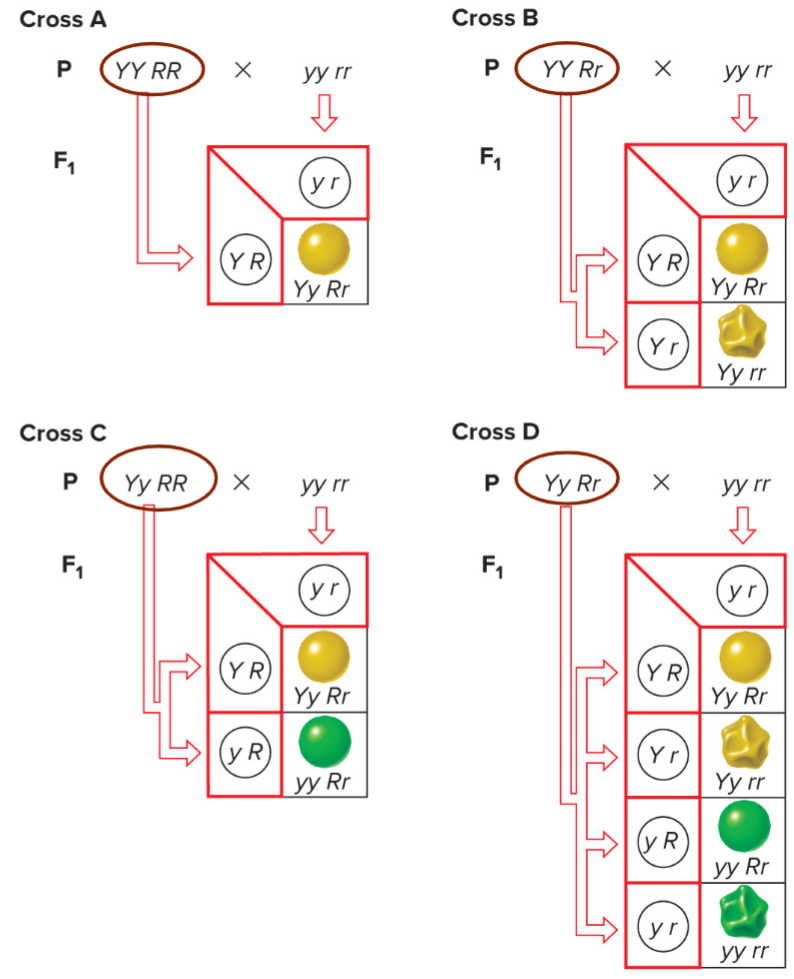
* very helpful for determining likelyhood for bigger punnet squares
Predicting proportions of progeny from multihybrid crosses example (lec 3)

Summary of Mendel’s 1865 paper (lec 3)
inheritance is particulate (not blended)
there are 2 copies of each trait in cell
gametes contain 1 copy of trait
alleles (diff form of trait) segregate randomly
alleles are dominant/recessive- thus diff btwn genotype and phenotype
diff traits assort indendently
Mendel’s traits are encoded in DNA (lec 3)
flow of genetic information in cells:
DNA to RNA to Protein
allelic diff at DNA lvl can influence mRNA expression and/or protein func, thus the phenotype
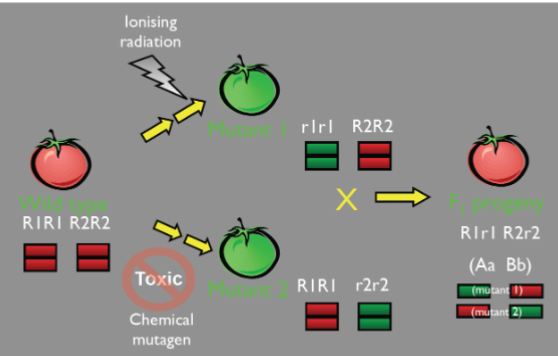
Mutations, source of allelic variation (lec 3)
mutation is the process where genes change from 1 allelic form to another, the creation of entirely new alleles can occur
genes mutate randomly, at any time and in any cell of an org
can arise spontaneously during normal replication/can be induced by a mutagen
only mutations in germline cells can be transmitted to progeny, somatic mutations can’t be transmitted
inherited mutations appear as alleles in populations
human genomes ~99.9% identical
most genetic variations created by SNPs (single nucleotide polymorphism)
Wild-type (+): most common allele (frequency >/= 1%)
mutant allele: rare allele (frequency < 1%)
mutations affecting phenotype occur very rarely
major cause of genetic diversity
can also be detrimental
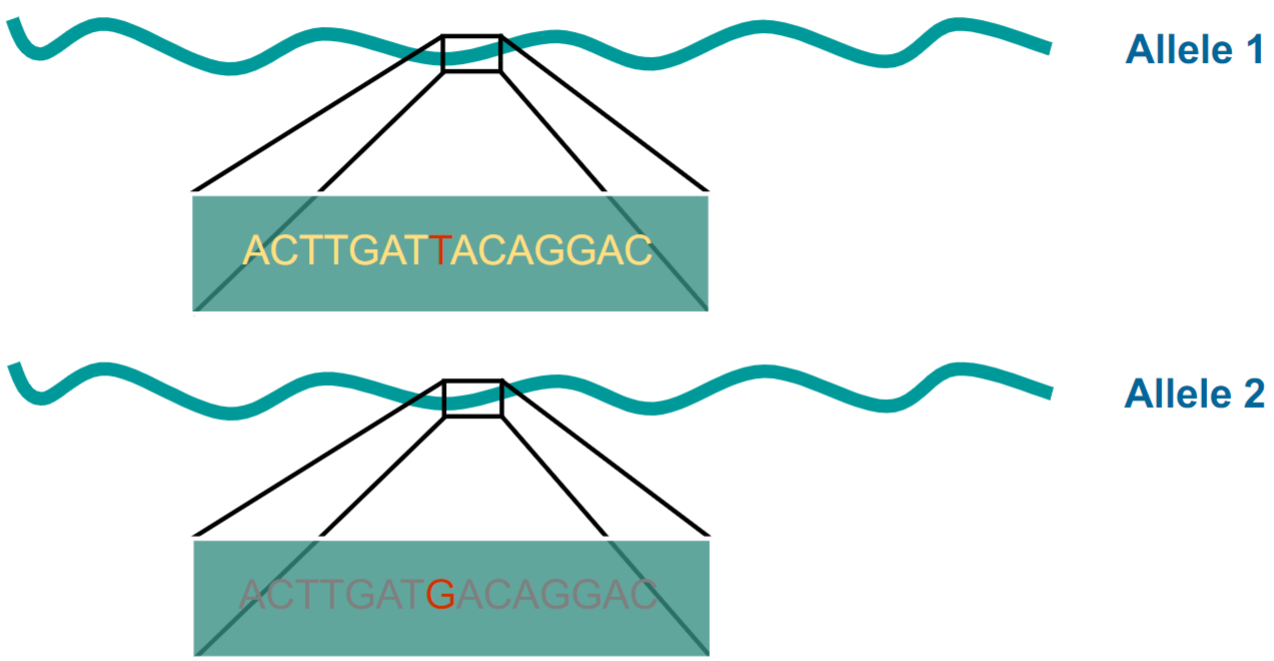
Single Nucleotide Polymorphisms (SNPs) are allelles (lec 3)
each SNP can be tracked back to genome change that occurred in single ancestral genome
Types of mutations (lec 3)
point mutations (SNPs):
ATGCAGT to ATCCAGT
Deletion/insertion (indel mutations):
Deletion: ATGCAGT to AT CAGT
insertion: ATGCAGT to ATGGCAGT
Change in repeat number:
CGGCGGCGG (3) to CGGCGGCGGCGGCGGCGG (6)
Chromosomal rearrangements:
Inversion: ATGCAGT to TGACGTA
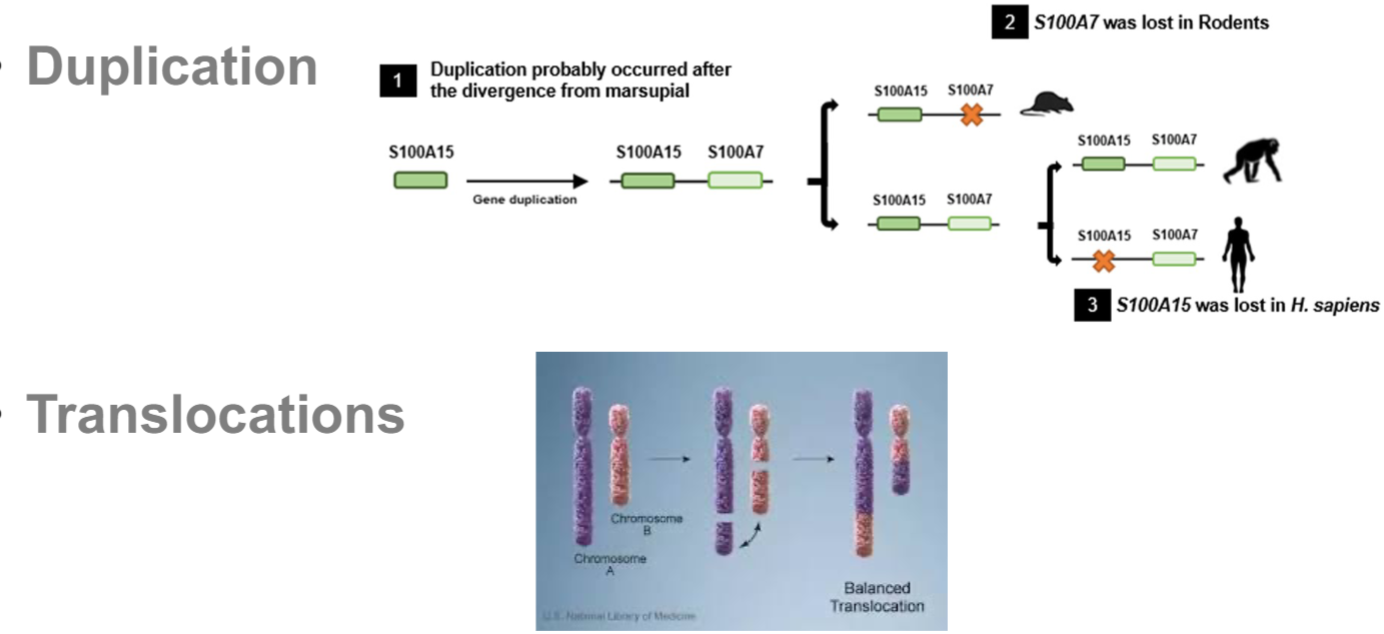
duplication: major drivers in evolution (extra copy of gene)
Translocations: exchange of genetic material btwn 2 diff chromosomes
Types of mutations in coding sequence of genes (lec 3)
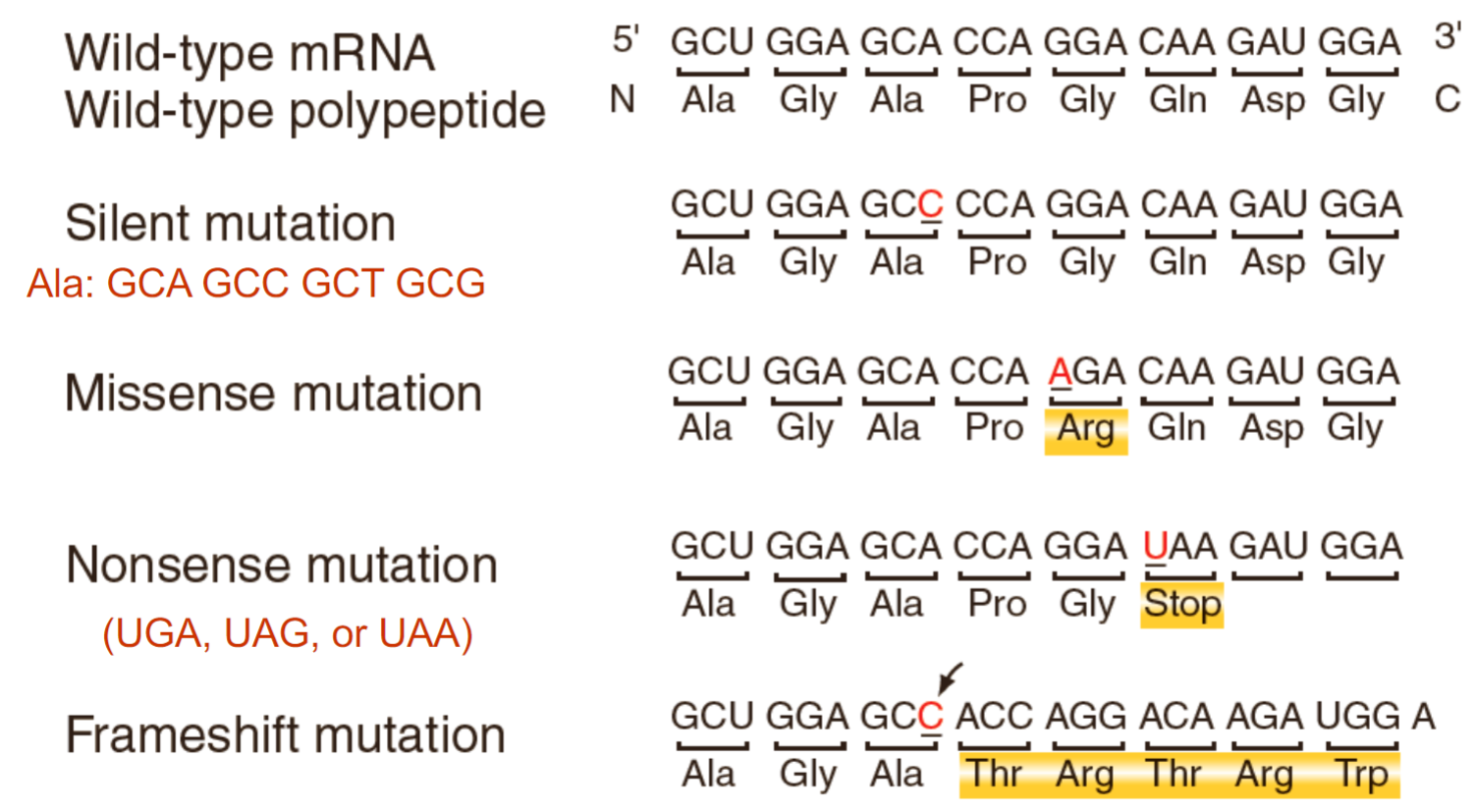
silent mutation: mutation that doesn’t change amino acid sequence
missense mutation: mutation that changes amino acid sequence (can be detrimental to protein depending on amino acid replaced)
nonsense mutation: mutation that causes there to be early stop codon (UGA, UAG, UAA)
frameshift: change in reading sequence (bc codons read in 3 nucelotides)
Gene expression and alleles (lec 3)
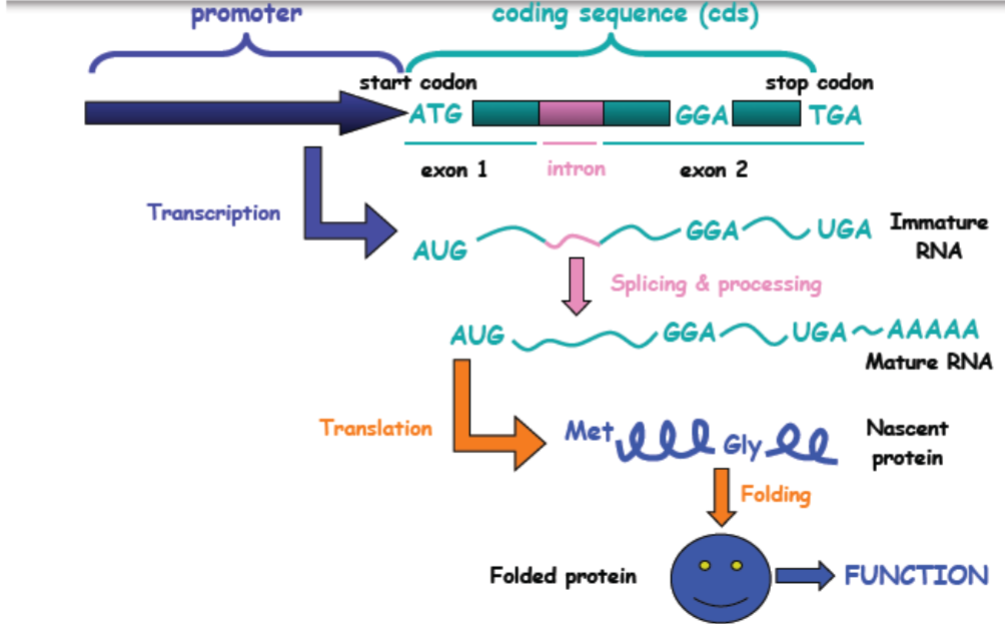
Gene expression and alleles: missense mutation (lec 3)
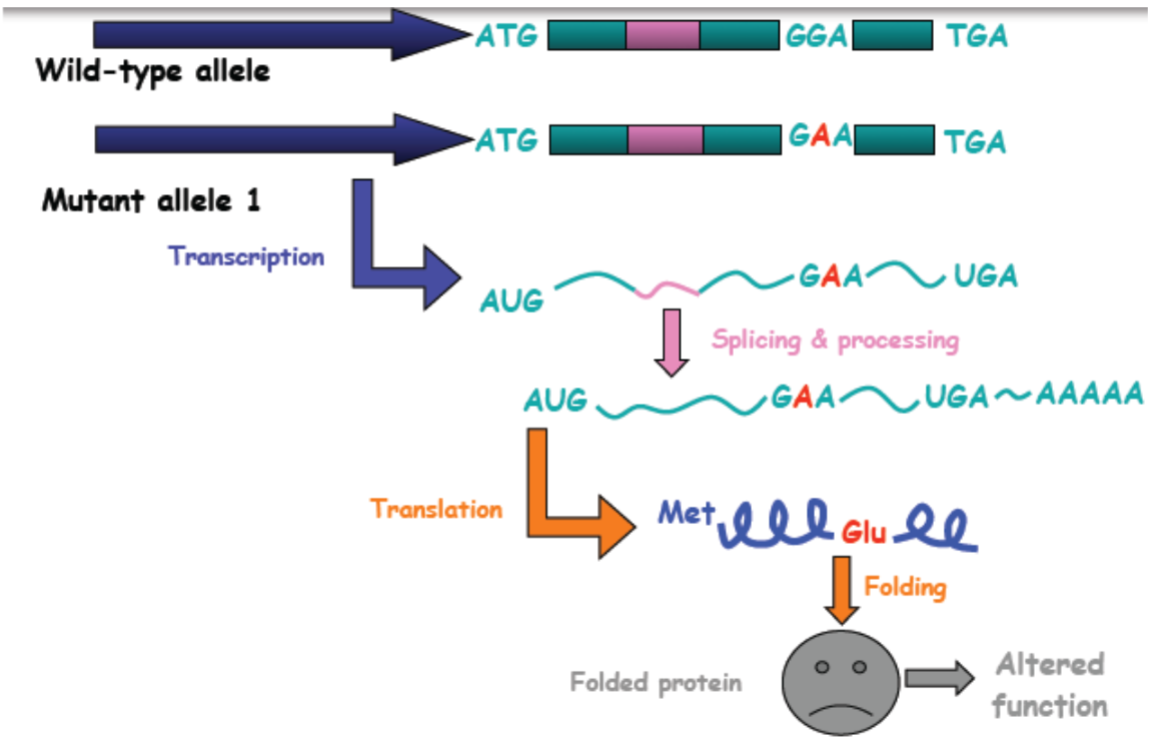
Gene expression and alleles: Splicing mutations (lec 3)
Splice donor/acceptor site mutations:
disrupts splice donor/acceptor site, resulting in incorrect retention/excision
often leads to large additions/deletions that may cause frameshifts
Leaky mutation: protein still functions but at worse level
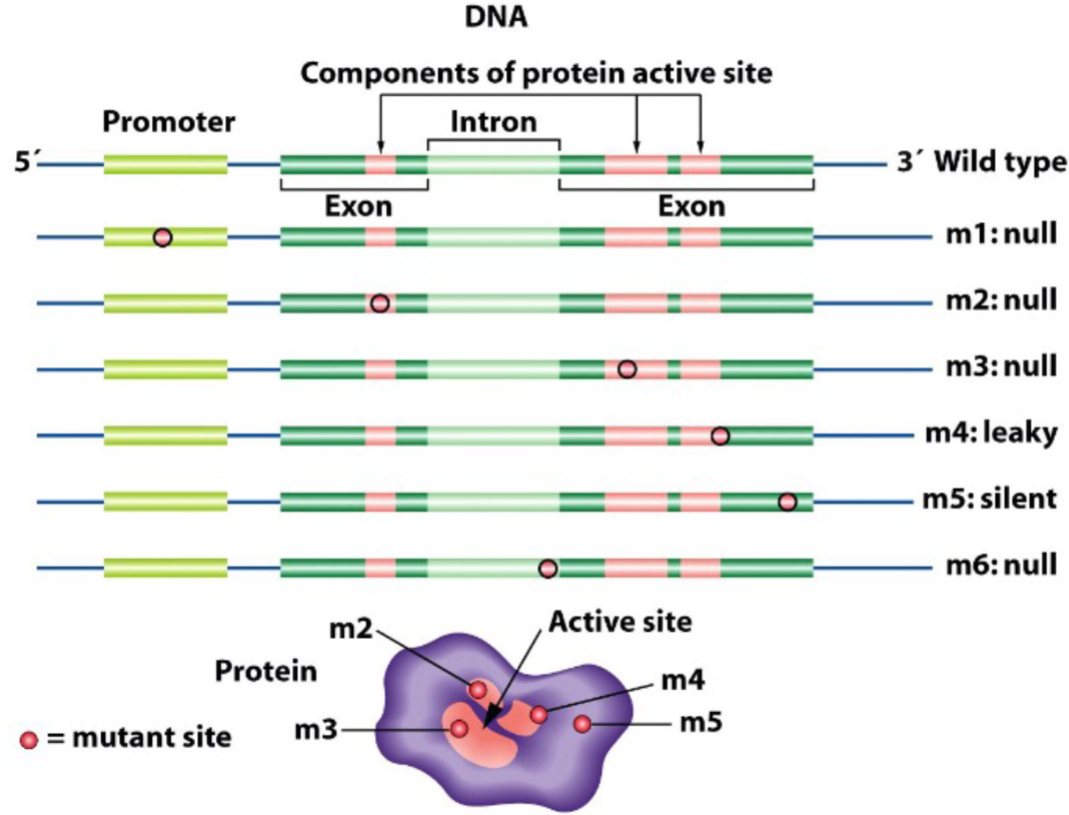
How mutations affect phenotype (lec 3)
think of central dogma of molecular bio (DNA to mRNA to Protein to organismal traits
allelic differences at DNA level can influence mRNA expression and/or protein function, thus affecting the phenotype
Genetic basis of single-gene disorders: PKU
PKU: phenylketonuria (chromosome 12)
missing enzyme leads to mental deficiency
in 1/10000 Caucasians
Phenylalanine (Phe, F) → Tyrosine (Tyr, Y)
without proper pH phenylalanine produces phenylpyruvic acid
build-up of phenylpyruvic acid can interfere w/ nervous system development
many possible sites for allelic diversity (PAH gene)
mutations in both exons + introns (interfering w/ splicing) can inactivate gene, causing PKU
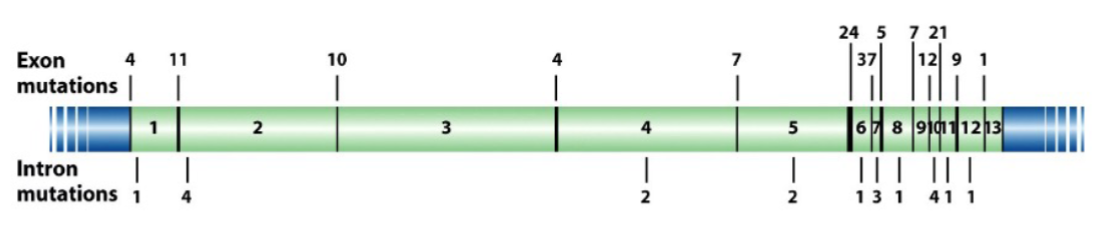
Gene basis of Mendel’s antagonistic pairs (lec 3)
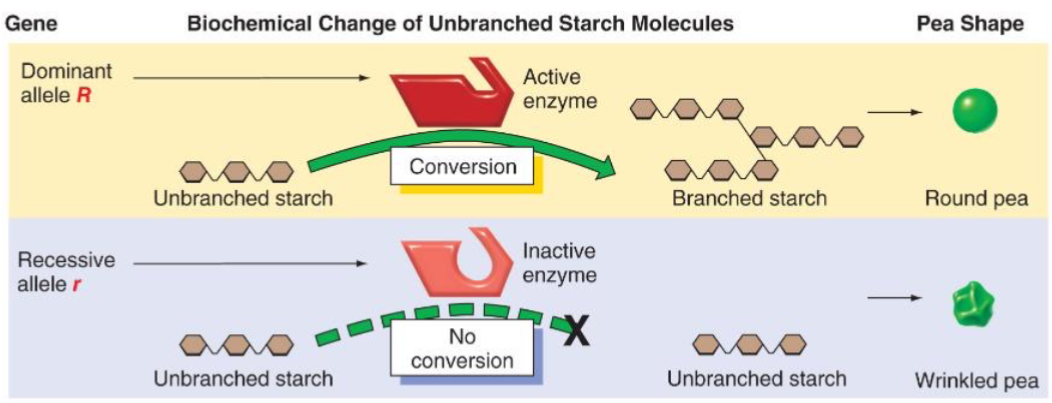
gene responsible for pea shape (Sbe 1)
Wrinkled allele: R disrupted by insertion to r
gene responsible for pea colour (Sgr1 YY)
functions in pathway involved breaking down chlorophyl during pea maturation = yellow peas
y allele is null allelle = no Sgr1 protein product = green peas
Loss of function mutations (lec 3)
result in reduced/abolished protein activity
usually recessive
Null (amorphic) mutations:
completely block function of gene product (ex: deletion of entire gene)
Hypomorphic mutations:
gene product has weak, but detectable activity
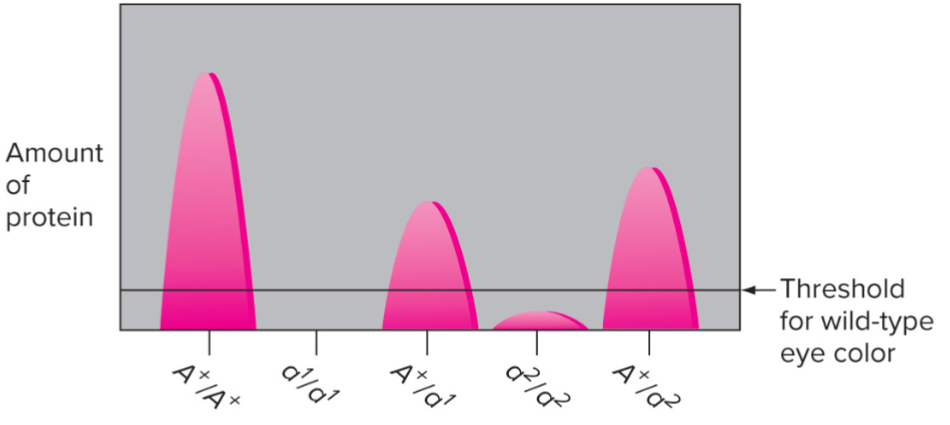
understanding recessiveness for ‘null’ alleles: for many genes, 50% of protein product is sufficient to give wild type phenotype
single WT (wild type) allele is Haplosufficient
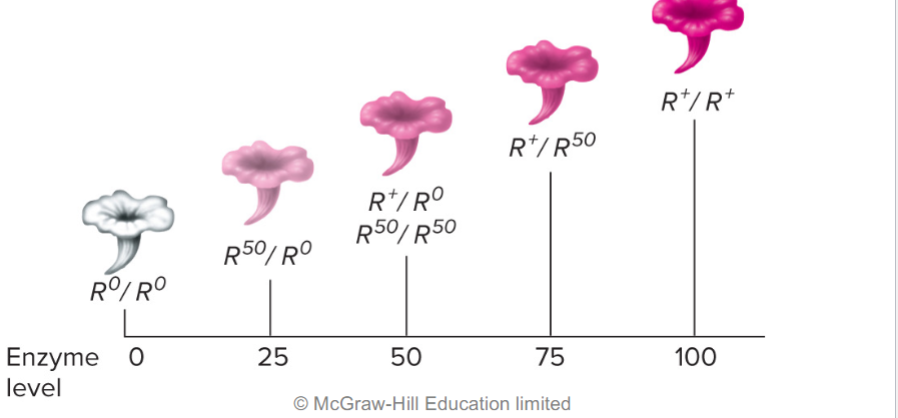
Incomplete dominance (lec 3)
phenotype varies w/ amount of functional gene product
ex: white and red rose, would lead to shades of pink when genes are mixed
Haploinsufficiency (lec 3)
one WT allele isn’t enough
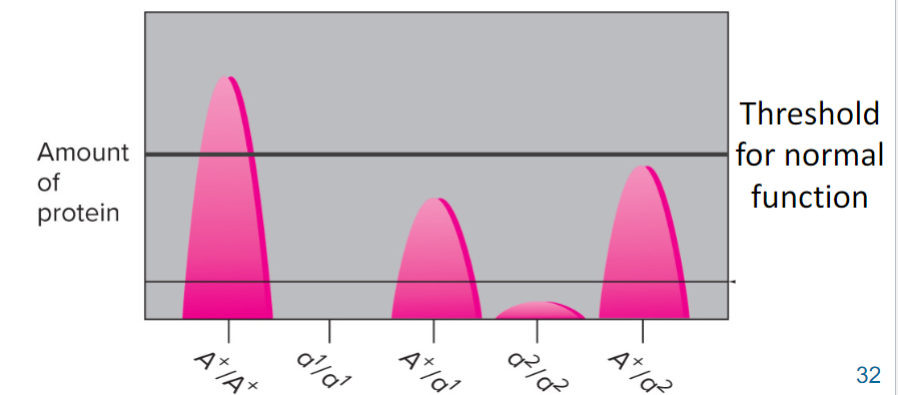
Some nonfunctional mutations can be dominant-negative (lec 3)
usually occurs in genes that encode multimeric proteins (made of >/= 2 subunits
mutant subunits block activity of normal subunits (mutant toxic to WT)
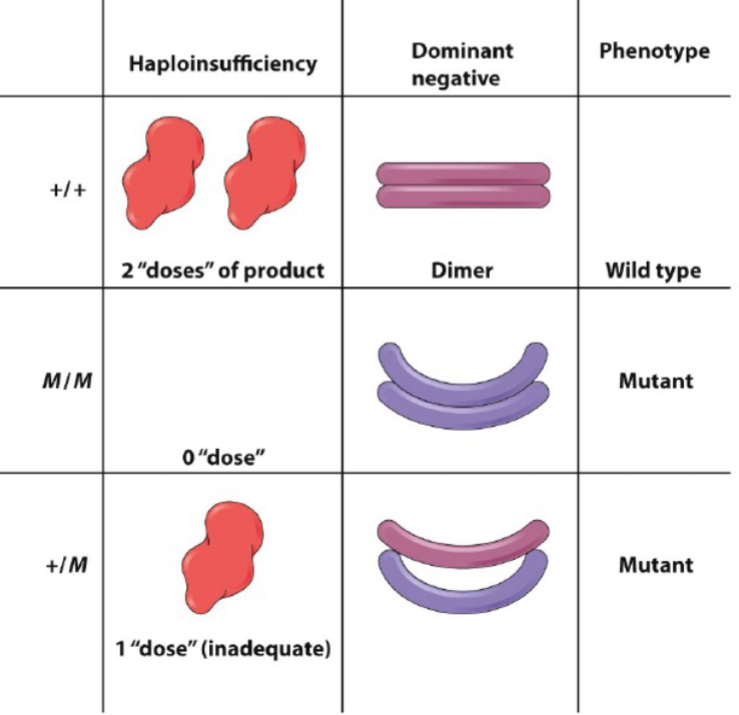
Gain of function mutations (lec 3)
enhance a function/confer new activity
usually dominant
Hypermorphic mutations:
generate more gene product/same amount of more efficient gene product
Neomorphic mutations:
generate gene product w/ new function/is expressed at inappropriate time/place
ex: antennapedia gene of fruit fly causes ectopic expression of leg determining gene in structures that normally produce antennae
Determining inheritance patterns in humans (lec 4)
tricky process due to:
long generation time
small numbers of progeny
no controlled mating
no pure-breeding lines
complex traits
Solutions to humans not being genetic models (lec 4)
use other organisms as models
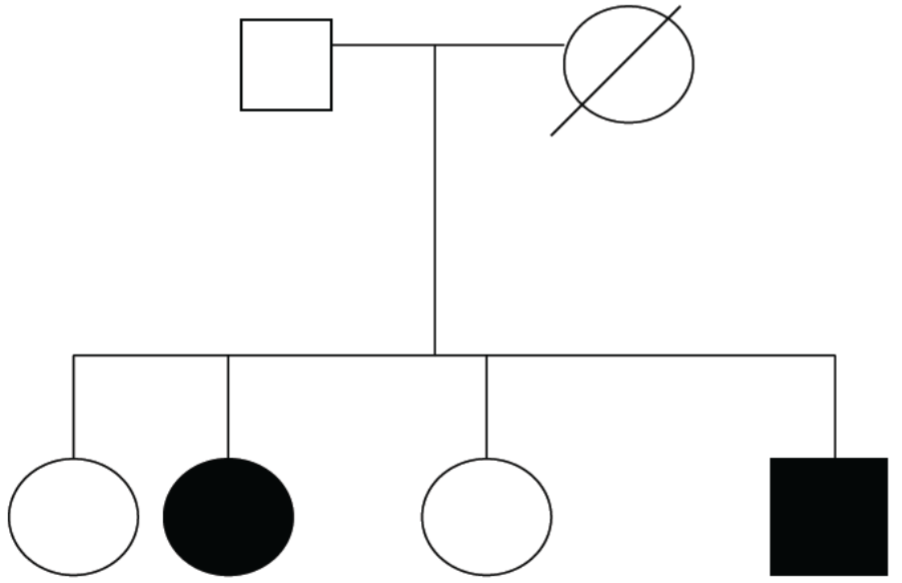
follow pedigrees: an orderly diagram of family’s relevant genetic features extending through multiple generations
pedigrees help us infer if trait is from single gene and if the trait is dominant/recessive
Diagram: shows disease is recessively inherited
Mendelian inheritance and humans: autosomal inheritance (lec 4)
humans autosomal traits are located on non-sex chromosomes (1-22)
may be inherited as:
autosomal dominant
autosomal recessive
Mendelian inheritance and humans: same principles apply (lec 4)
thousand of examples are described in database
idea of dominant/recessive traits
Mendelian inheritance and humans: autosomal inheritance (dominant) (lec 4)
autosomal dominant:
homozygous dominant and heterozygotes exhibit affected phenotype
males and females are equally affected and may transmit the trait
affected phenotype doesn’t skip a generation
Mendelian inheritance and humans: autosomal inheritance (recessive) (lec 4)
autosomal recessive:
only homozygous recessive individuals exhibit affected phenotype
males and females are equally affected and may transmit the trait
may skip generations
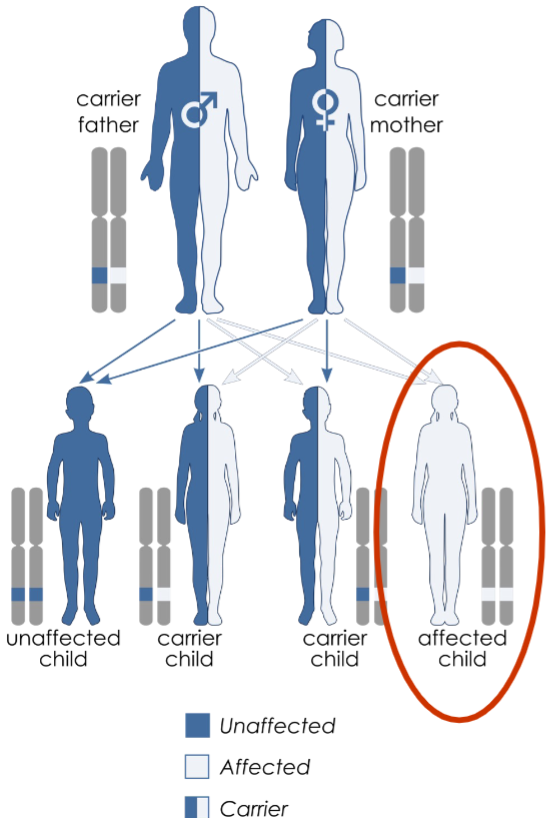
Anatomy of pedigree (lec 4)
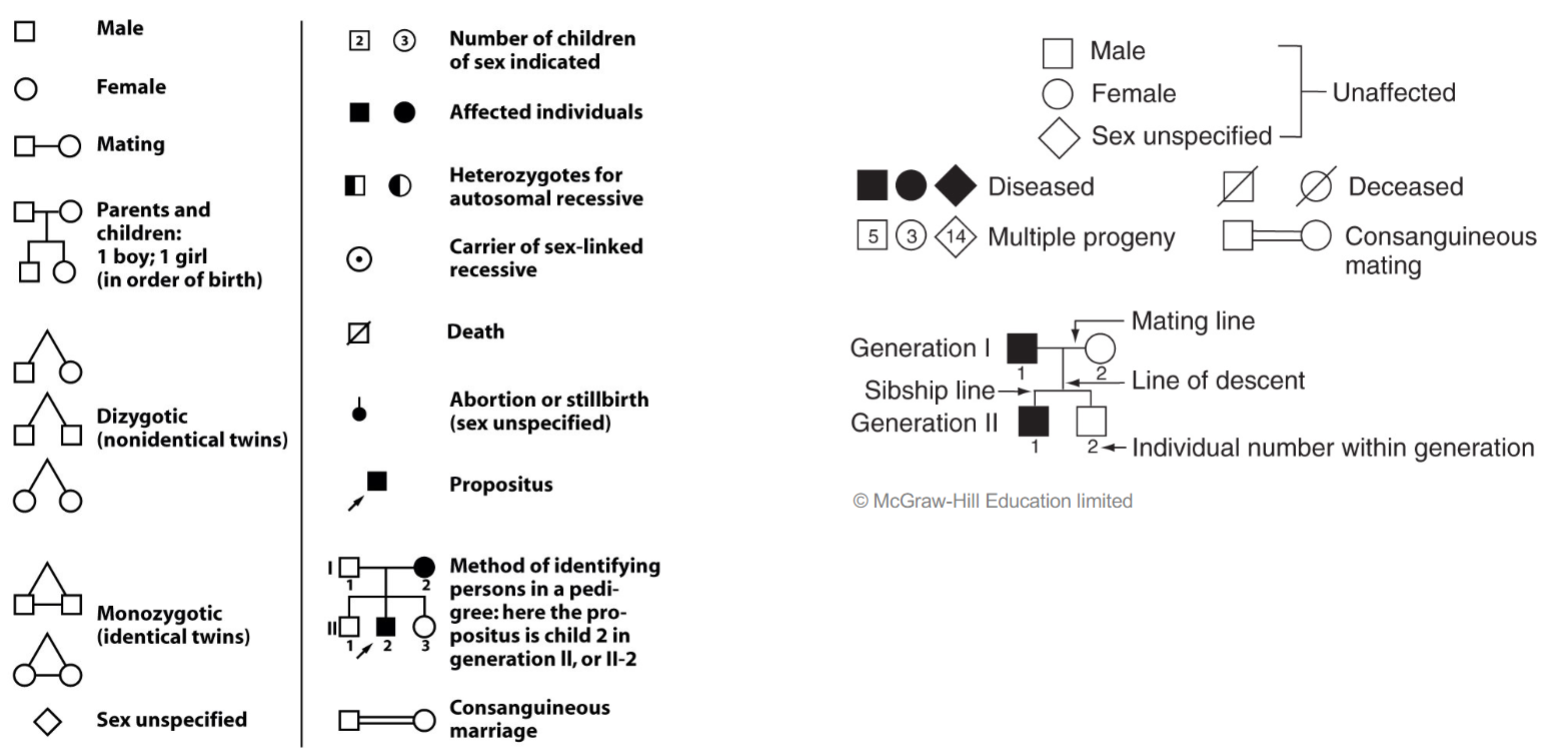
Anatomy of pedigree: Huntington’s disease (lec 4)
vertical pattern of inheritance indicates rare dominant trait
alters human neurodevelopment
people who carry mutation can live healthy lives for 4+ decades(~40) before onset of symptoms
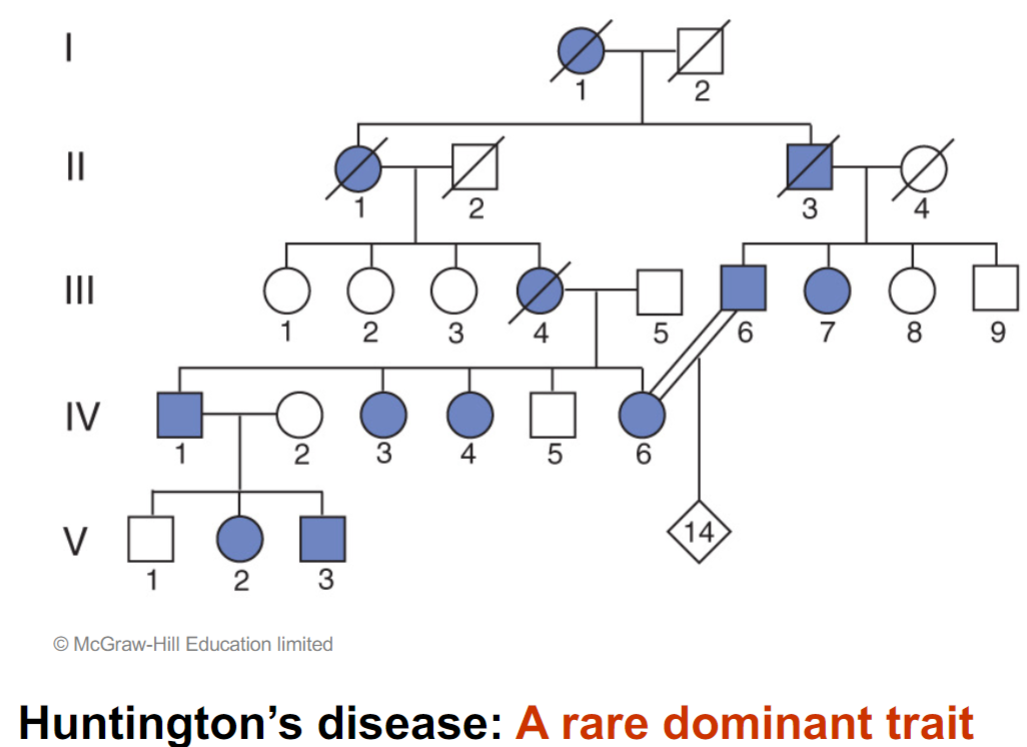
Mendelian inheritance of Huntington’s disease (lec 4)
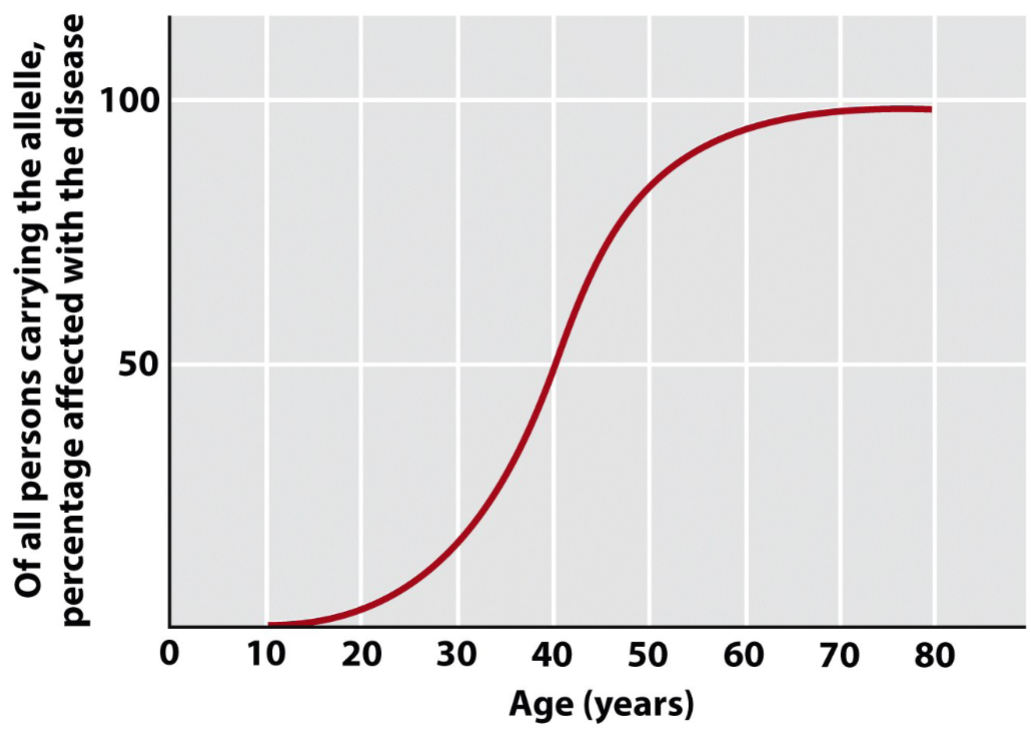
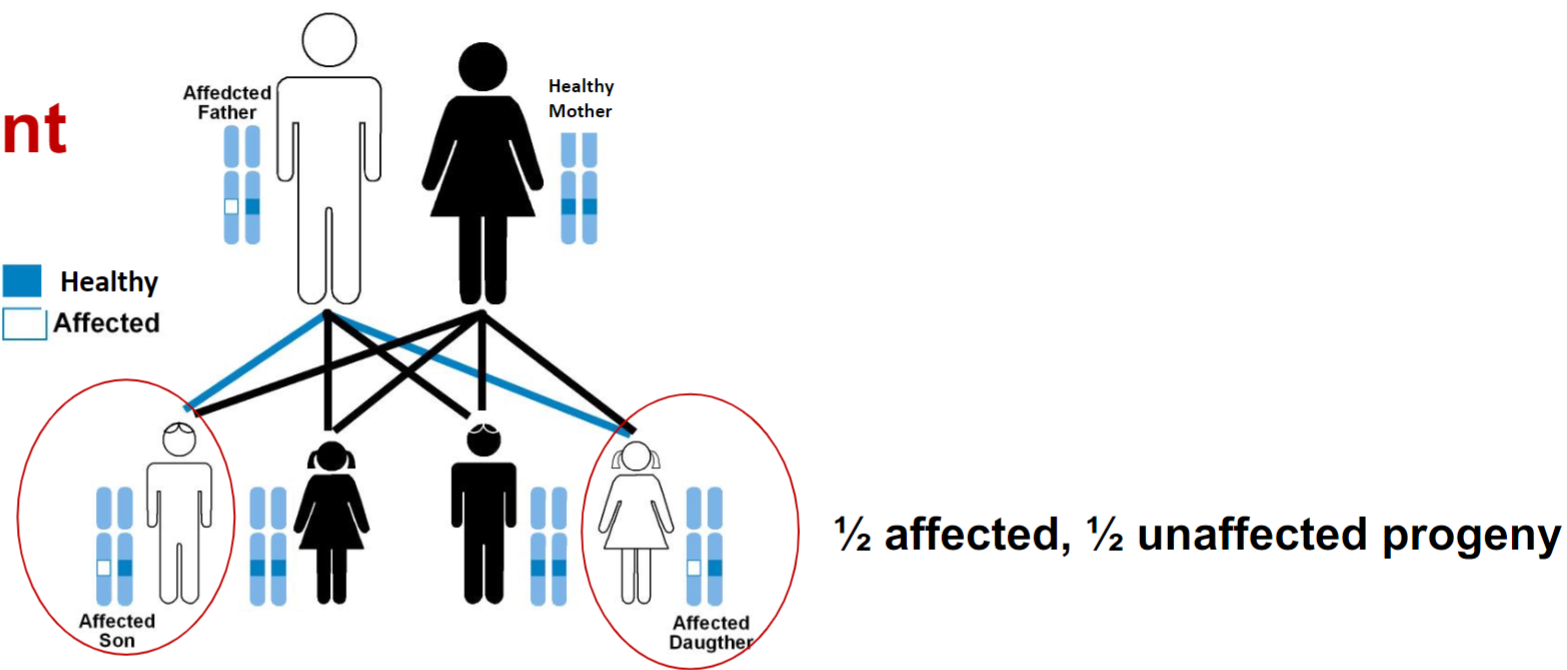
Recognizing dominant traits in pedigrees (lec 4)
3 key aspects of pedigrees w/ dominant traits:
affected children always have at least one affected parent
as a result, dominant traits show a vertical pattern of inheritance: trait shows up in every generation
2 affected parents can produce unaffected children, if both parents are heterozygotes
Horizontal pattern of inheritance (lec 4)
indicates rare recessive trait
parents are unaffected but are heterozygous (carries) for recessive allele
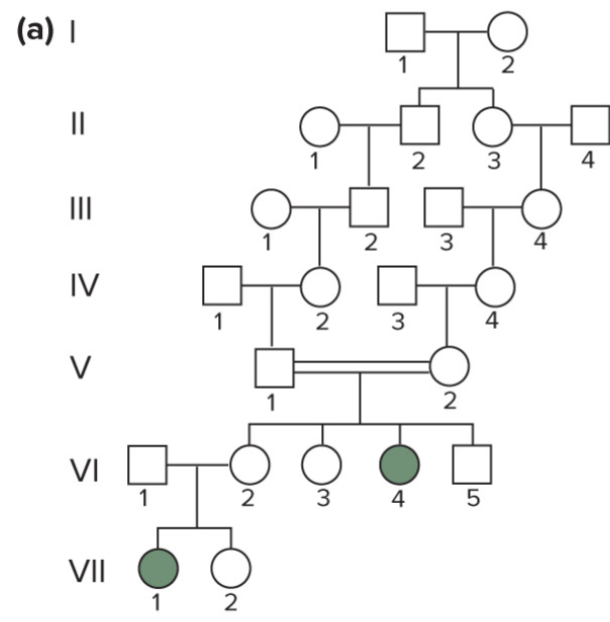
Recognizing recessive traits in pedigrees (lec 4)
4 key aspects of pedigrees w/ recessive traits:
affected individuals can be children of 2 unaffected carriers, particularly as the result of consanguineous matings
all children of 2 affected parents should be affected
rare recessive traits show horizontal pattern of inheritance: trait first appears among several members of one generation and is not seen in earlier generations
recessive traits may show vertical pattern of inheritance if trait is extremely common in the population
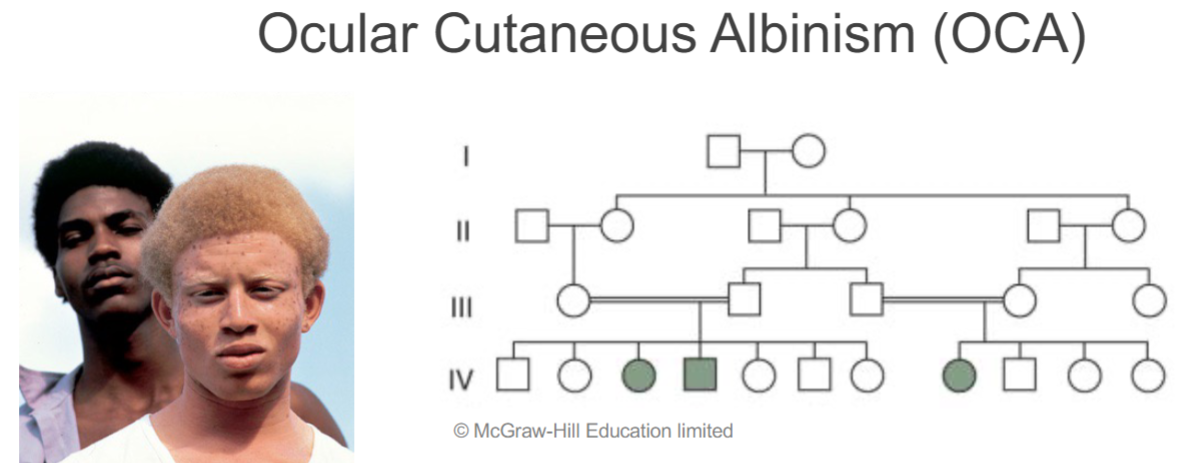
Consanguineous mating (lec 4)
pedigree w/ Consanguinity (inbreeding) frequently “uncovers” traits that are recessive
can give rise to “inbreeding depression” (offspring that are less fit than their parents)
Solving genetics problems (lec 4)
list genotypes and phenotypes for the trait
determine the genotypes of parents
determine parents’ possible gametes
determine possible genotypes of offspring
repeat for successive generations
is the trait rare/common in population?
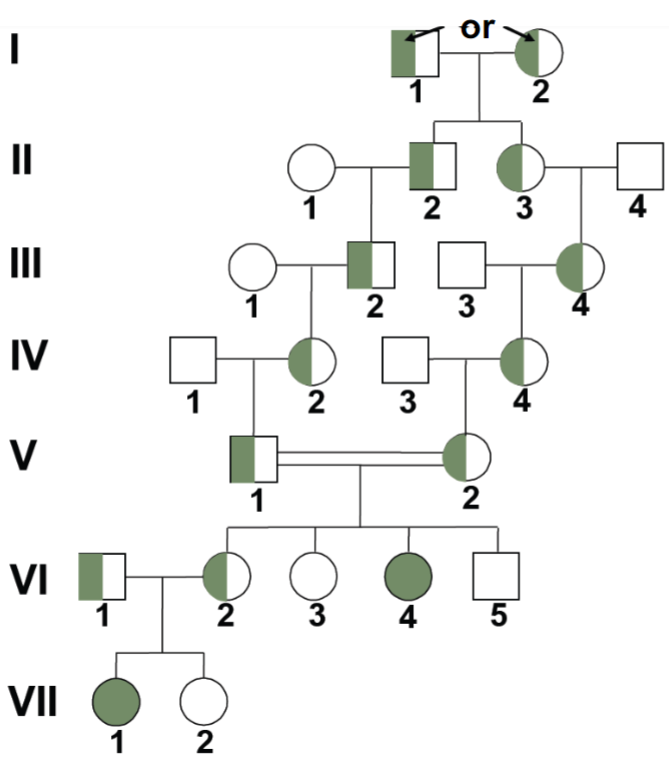
Solving pedigrees: deducing mode of inheritance and associated genotypes question 1 (lec 4)
what is known at the level of the phenotype: affected individuals coloured in
recessive trait bc skips generations
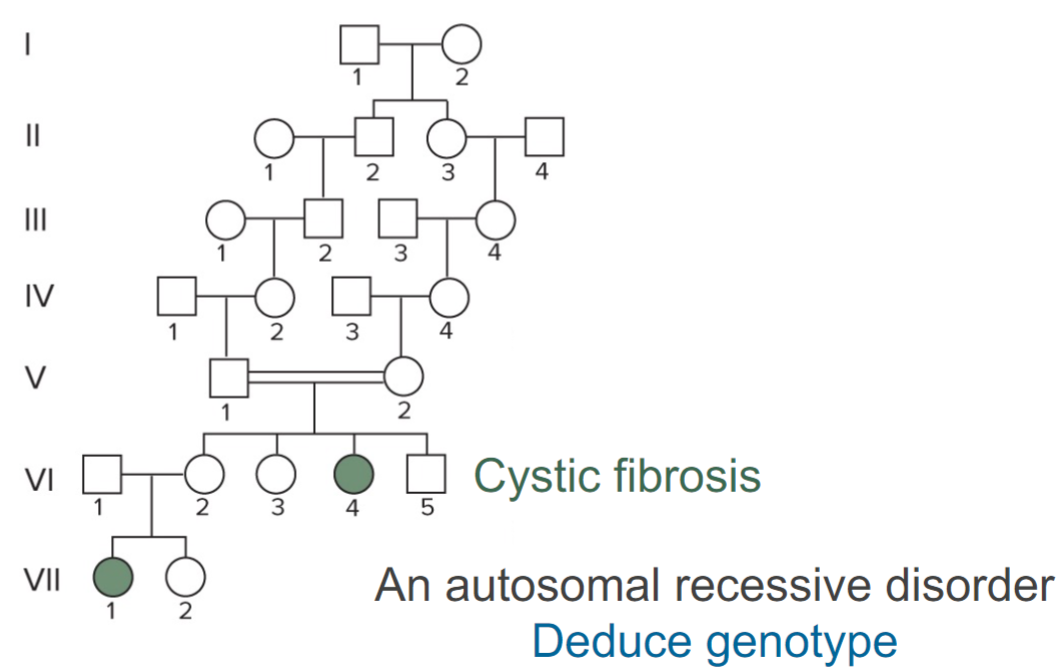
Solving pedigrees: deducing mode of inheritance and associated genotypes question 1 (lec 4)
parents of gen V and VI should be heterozygous bc has affected child but also unaffected children
Solving pedigrees: deducing mode of inheritance and associated genotypes question 1 (lec 4)
carriers of disease should be found in family because trait is rare
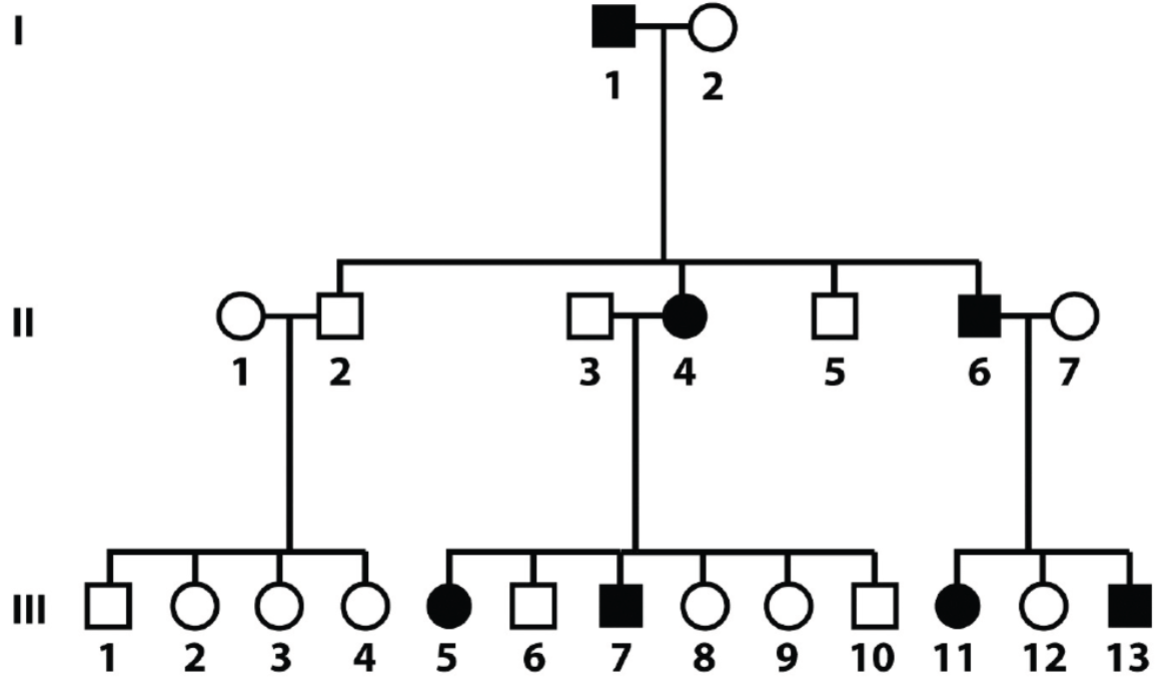
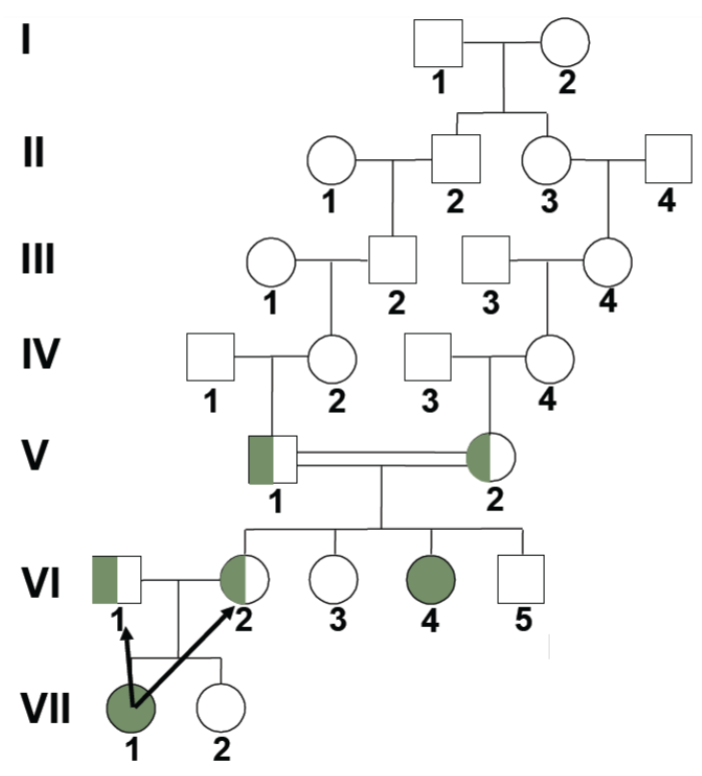
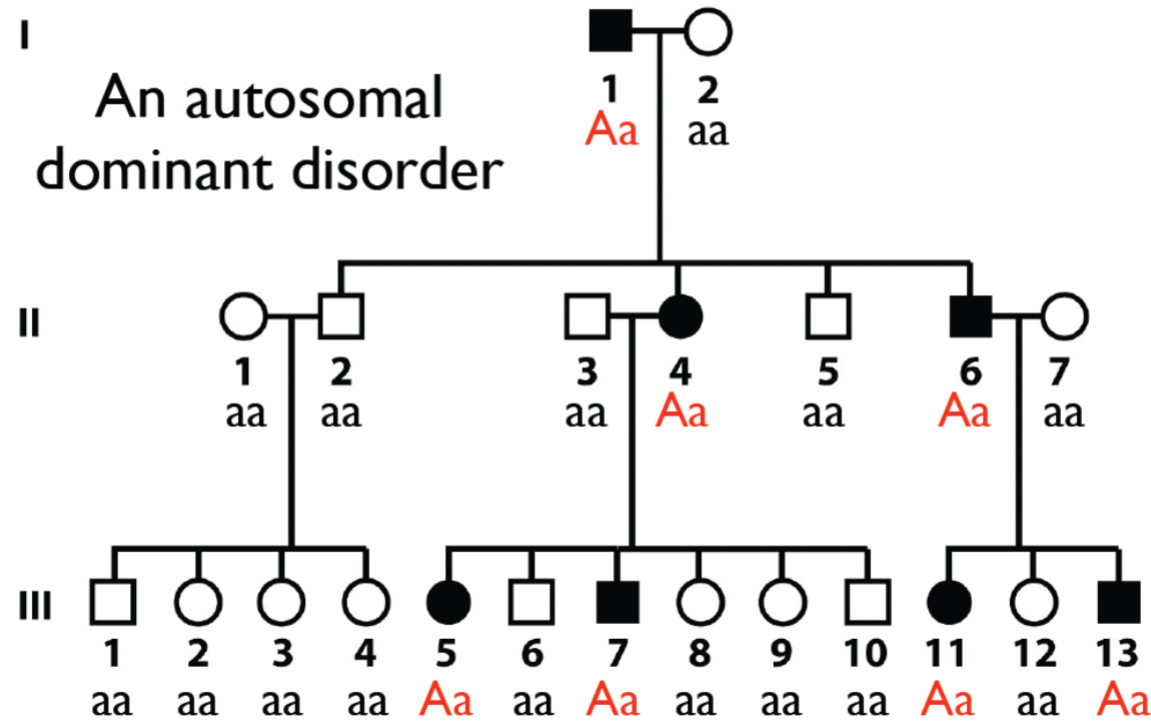
Solving pedigrees: deducing mode of inheritance and associated genotypes question 2 (lec 4)
shown is autosomal dominant bc disease shows up in every gen
therefore, if not showing disease means individual is homo recessive
disease individuals are hetero bc disease is dominant, if diseased was homo dominant, then all children should be affected
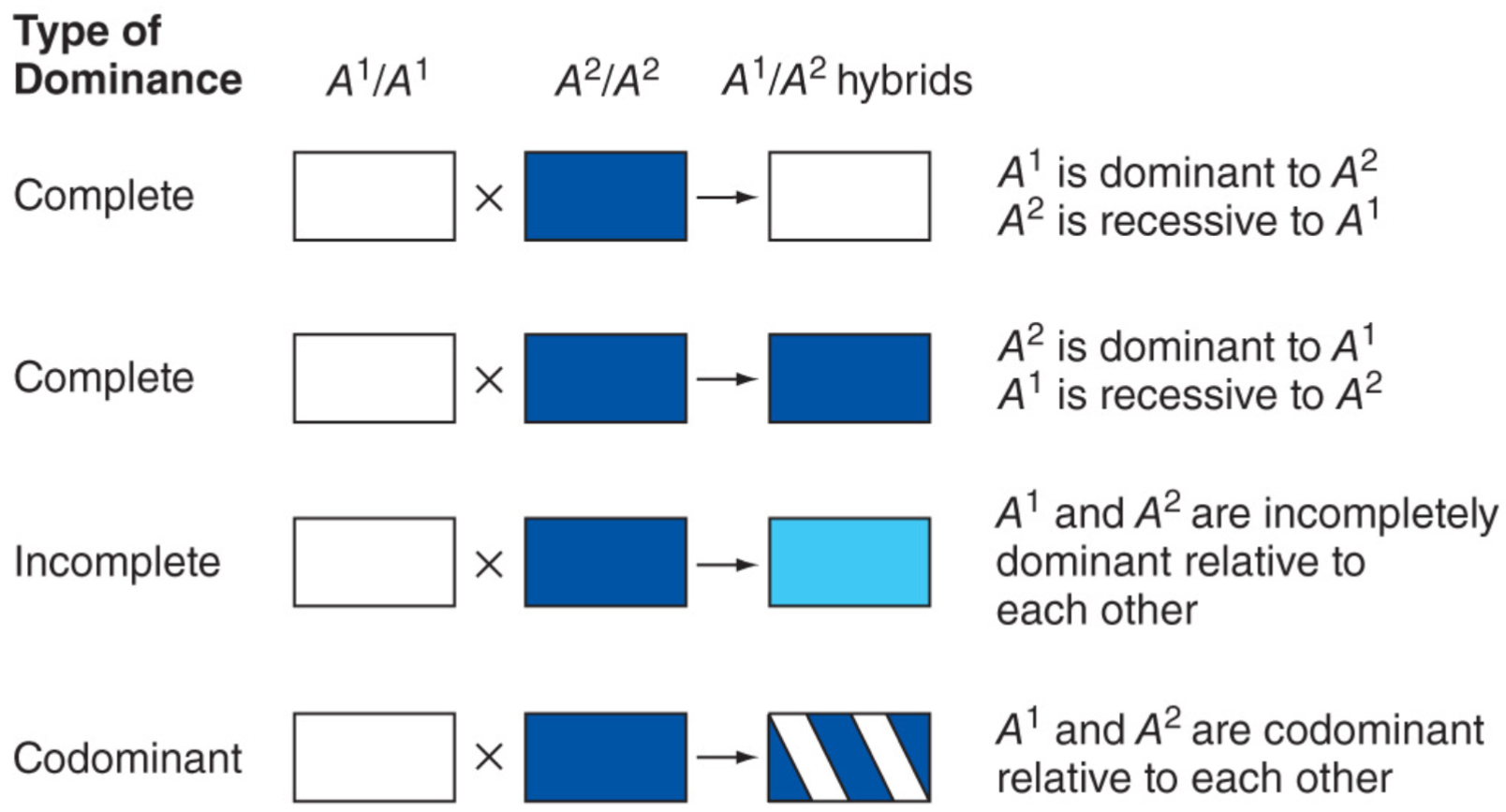
Reaction to mendel: Two Camps (lec 5)
exception to Mendel’s rules show that they aren’t general (some form of blending inheritance applies)
Try to explain exceptions in Mendelian terms of segregation and independent assortment
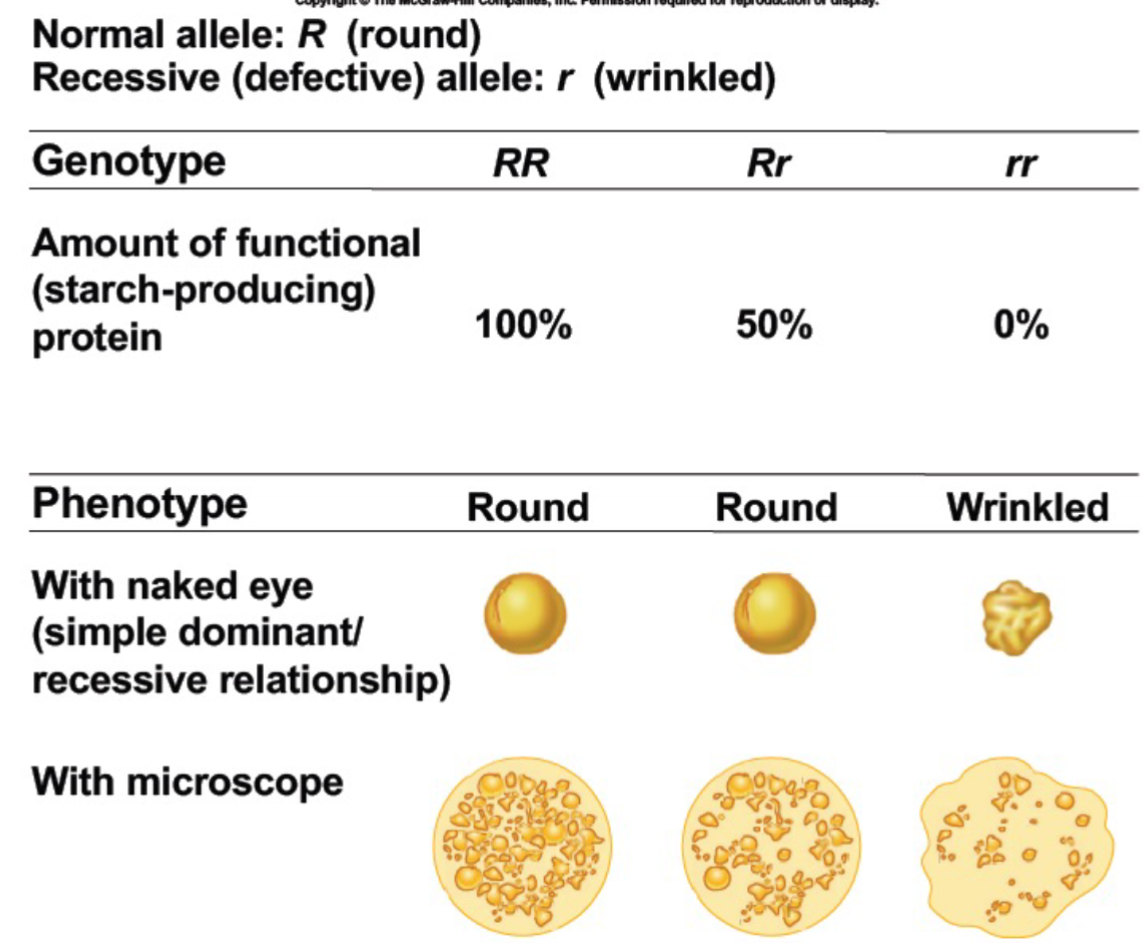
Dominance isn’t always complete (lec 5)
phenotype of heterozygote defines dominance relationship of 2 alleles
Complete dominance: only one trait shows (blue or white)
Incomplete dominance: mix of 2 traits show (white and blue mix to form sky blue)
Codominance: both traits show (white and blue form blue w/ white dots/stripes)
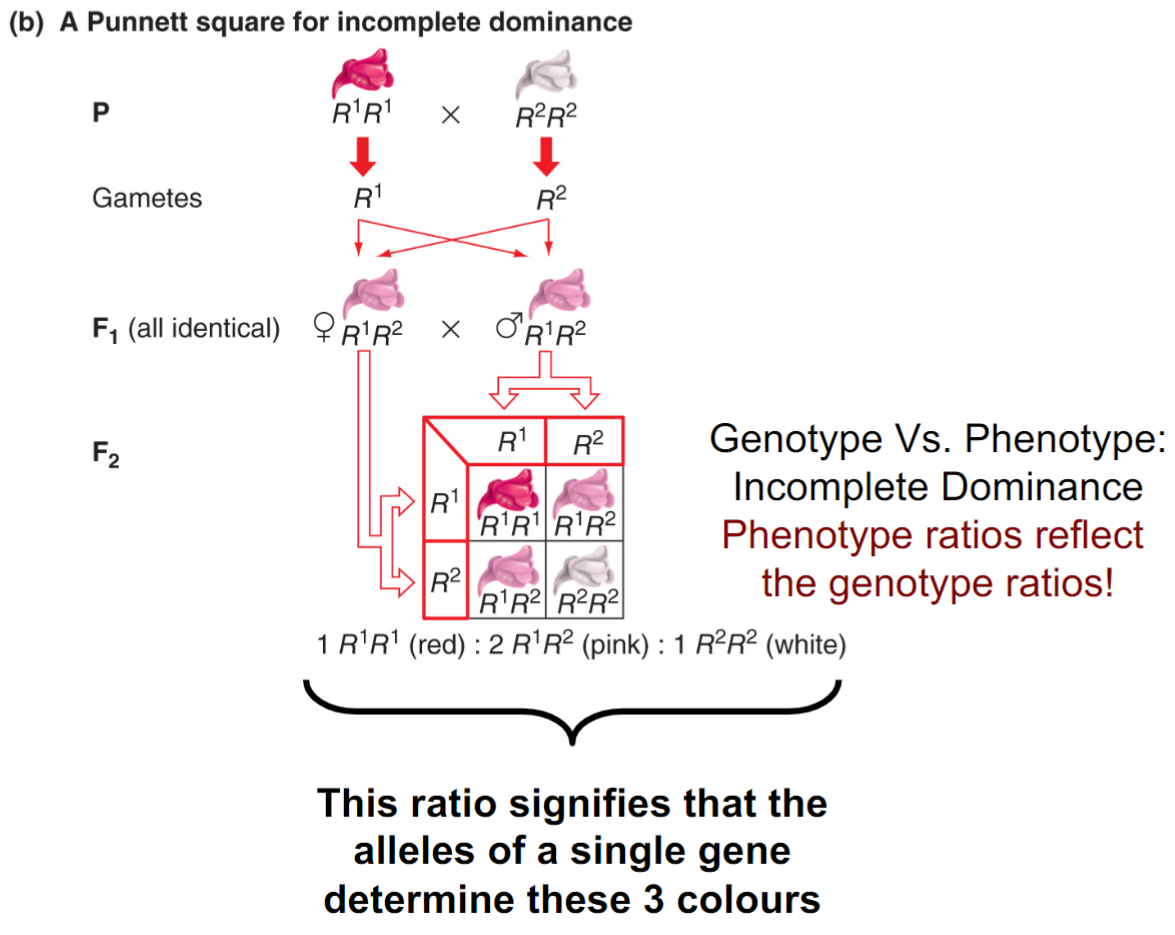
Exceptions to 3:1 ratio: incomplete dominance in Antirrhinum (lec 5)
crosses of pure-breeding red w/ pure-breeding white results in all pink F1 progeny (snapdragon flowers)
phenotypes reflect genotypic ratios
ratio signifies alleles of single gene determine these 3 colours (just a mix up of 3:1 shows monohybrid cross)
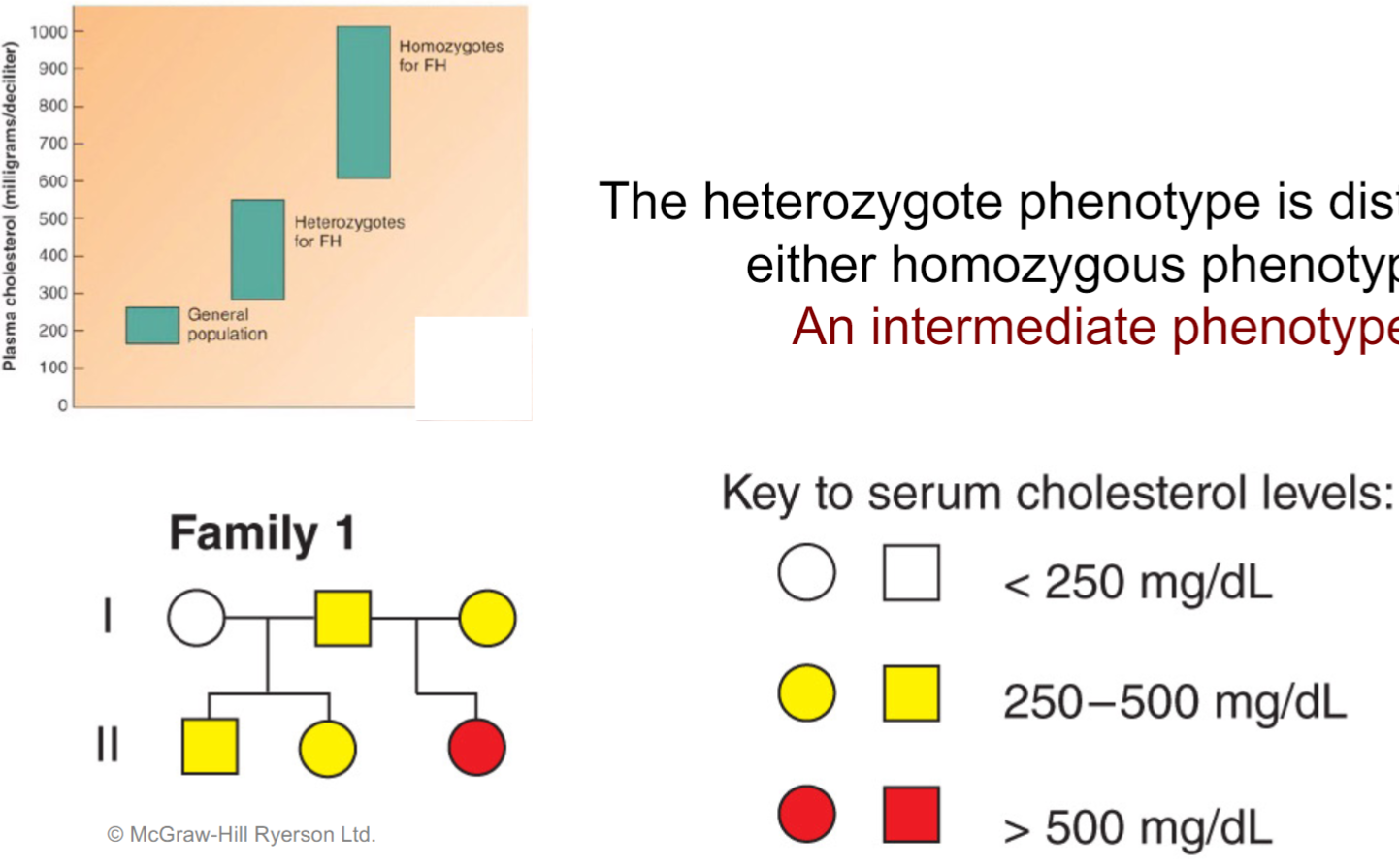
Incomplete dominance - familial hypercholesteraemia (lec 5)
heterozygote phenotype is distinct from either homozygous phenotype (intermediate phenotype)
Dominance is not always complete: summary (lec 5)
crosses btwn true-breeding strains can produce hybrids w/ phenotypes diff from both parents
Incomplete dominance:
F1 hybrids that differ from both parents express an intermediate phenotype. neither allele is dominant nor recessive to other
phenotypic ratios are same as genotypic ratios
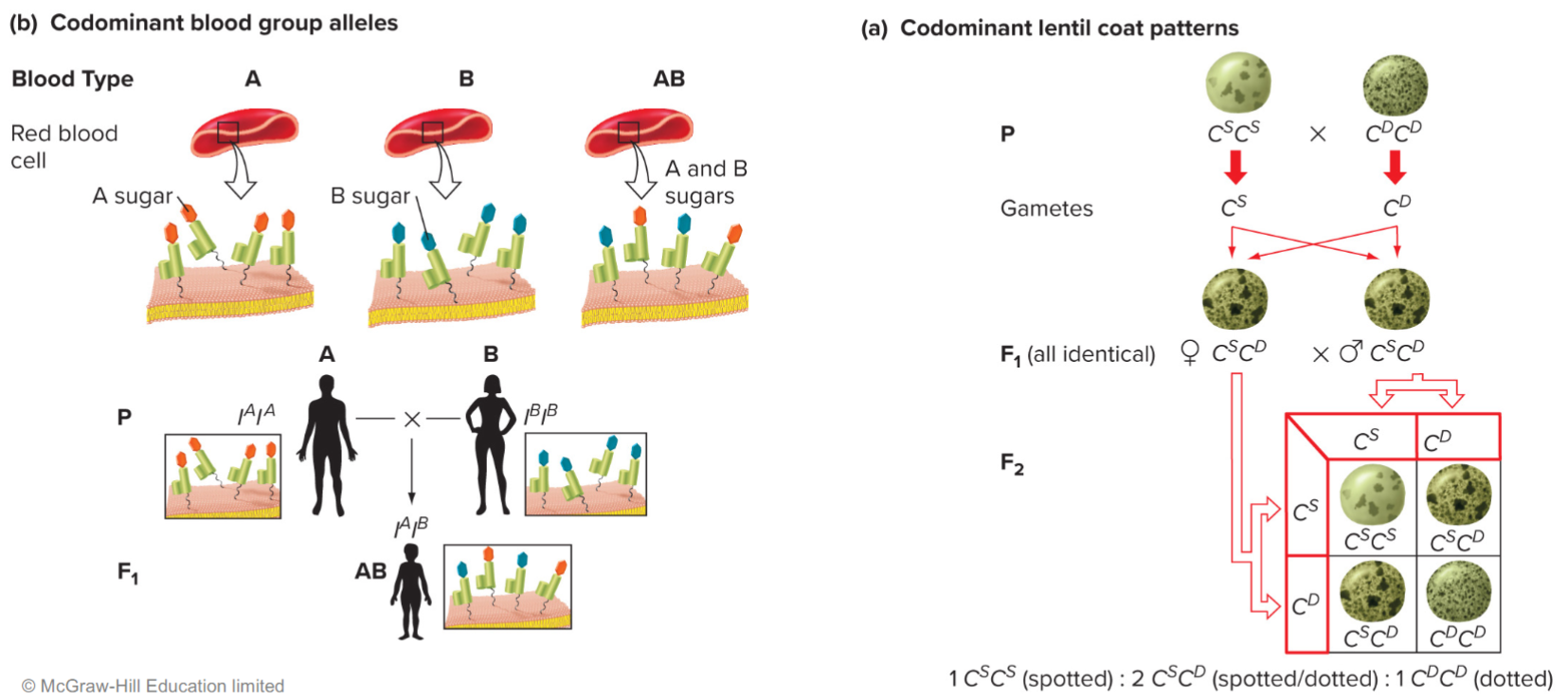
Codominance:
F1 hybrids express phenotype of both parents equally
phenotypic ratios are same as genotypic ratios
Co-dominant blood group and lentil coat pattern alleles (lec 5)
phenotype ratios reflect genotype ratios
Genes can have more than 2 alleles (lec 5)
multiple alleles of a gene can segregate in populations
although there may be many alleles in pop, each individual can carry only 2 of the alternatives:
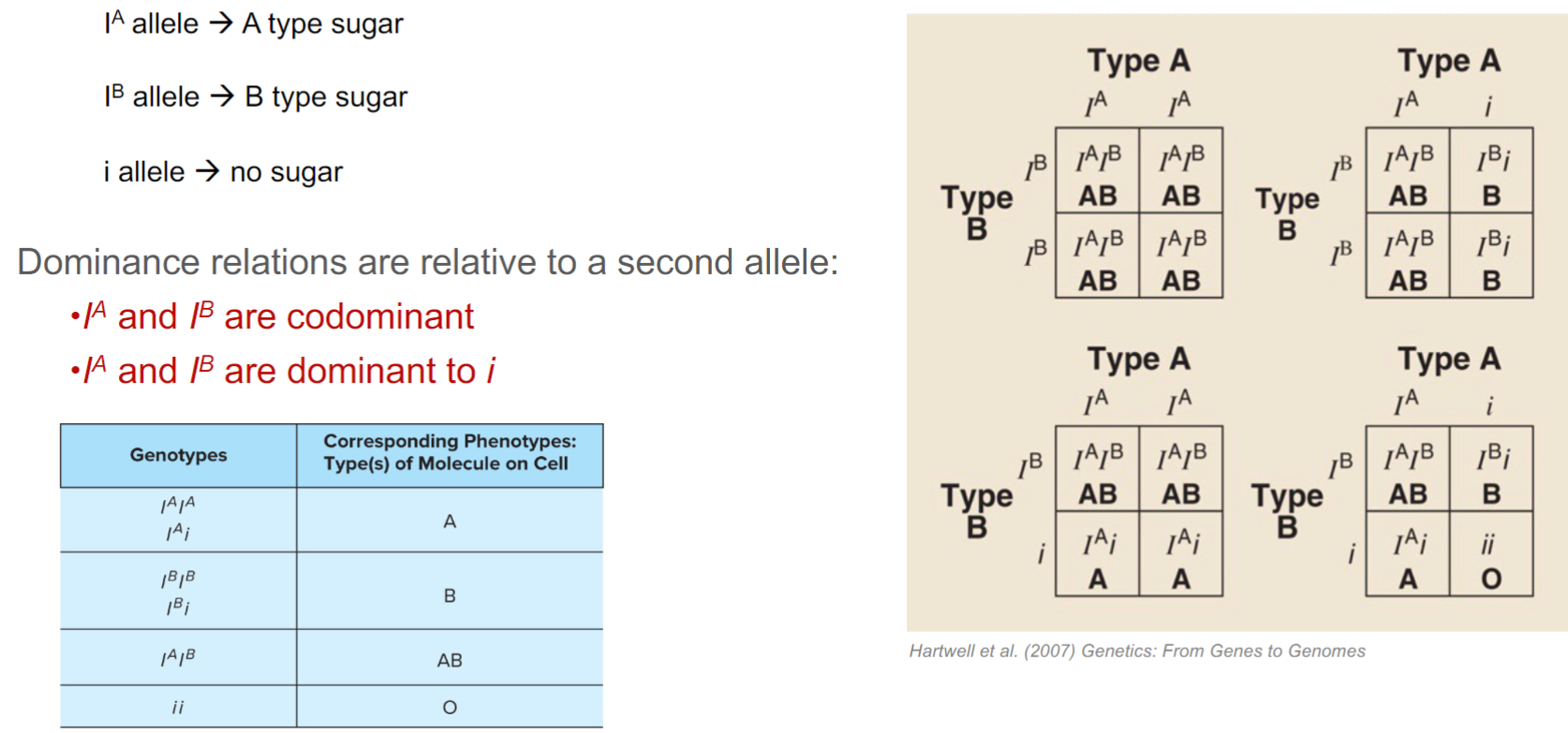
ABO blood group gene: I
3 alleles: IA, IB, and i
6 possible ABO genotypes: IAIA, IBIB, IAIB, IAi, IBi, or ii
Dominance relations are unique to pair of alleles:
dominance relations are always relative to second allele and are unique to a pair of alleles alleles
ABO blood group:
IA/B is completely dominant to i but codominant to IB/A
6 genotypes generate 4 phenotypes: Type A/B/AB/O
ABO blood types in humans are determined by 3 alleles of one gene (lec 5)
i is a null mutation because there is no sugar produced
Seed coat patterns in lentils are determined by a gene w/ five alleles (lec 5)
Five alleles for C gene: spotted (CS), dotted (CD), clear (CC), marbled-1 (CM1), and marbled-2 (CM2)
reciprocal crosses btwn pairs of pure-breeding lines is used to determine dominance relations
a 3:1 ratio in each cross indicates that diff alleles of same gene are involved

How do we establish dominance relations btwn multiple alleles of gene (lec 5)
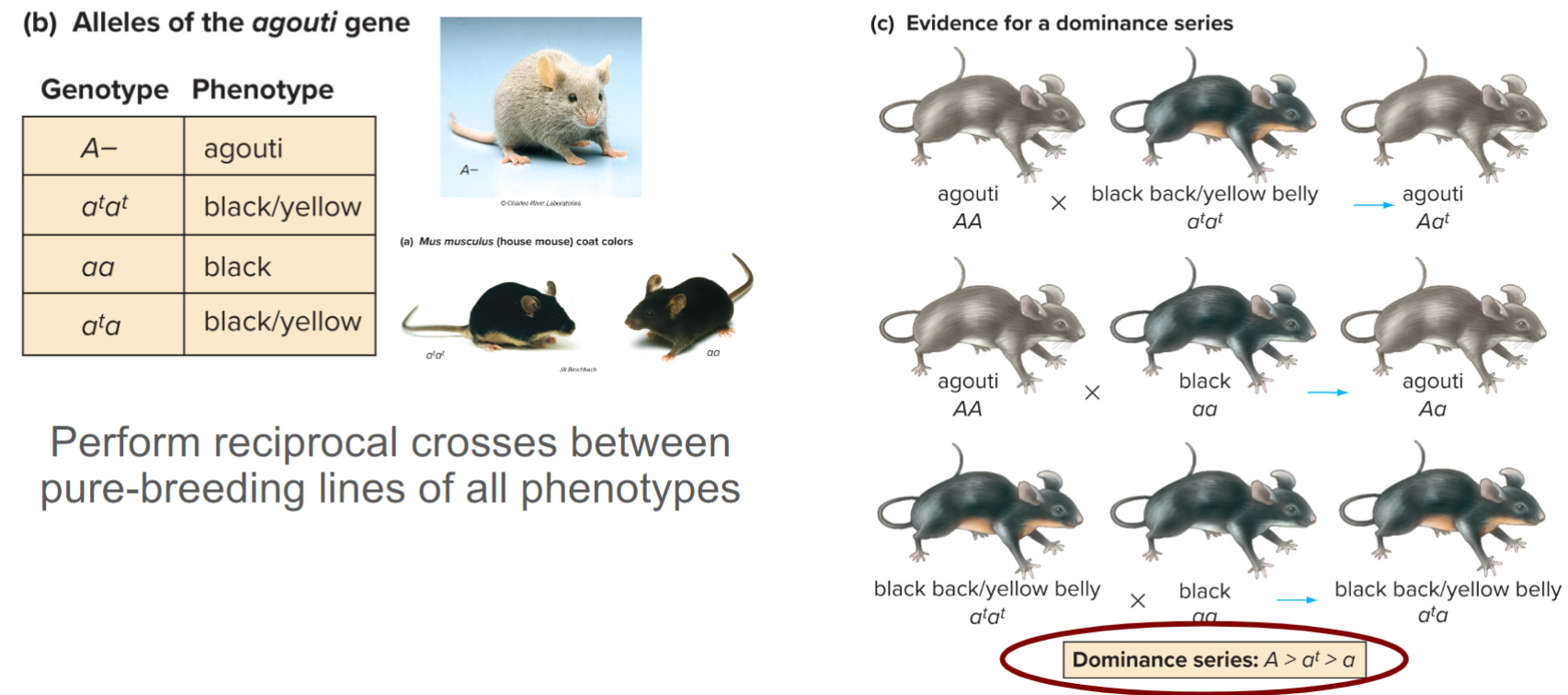
Dominance relations btwn alleles don’t affect transmission of alleles (lec 5)
type of dominance depends on type of proteins encoded and by the biochem func of proteins
Variation in dominance relations don’t negate Mendel’s law of segregation
Alleles still segregate randomly
Interpretation of phenotype/genotype relations is more complex
one gene could affect multiple traits (lec 5)
Pleiotropy: phenomenon of single gene determining several distinct and seemingly unrelated characteristics
EX: many aboriginal Maori men have respiratory problems and are sterile due to mutations in gene required for functions of cilia (failure to clear lungs) and flagella (immotile sperm)
with some pleiotropic genes:
heterozygote can have visible phenotype
homozygotes may be inviable (causes lethality)
alleles that affect viability often produce deviations from 1:2:1 genotypic and 3:1 phenotypic ratio predicted by Mendel’s laws
Lethality and Pleiotropy (lec 5)
AY is dominant to A for hair colour but recessive to A for lethality
cross yellow x yellow mice
F1 mice are 2/3 yellow and 1/3 agouti
2:1 ratio is indicative of a recessive lethal allele
Pure-breeding yellow (AYAY) mice can’t be obtained because they aren’t viable
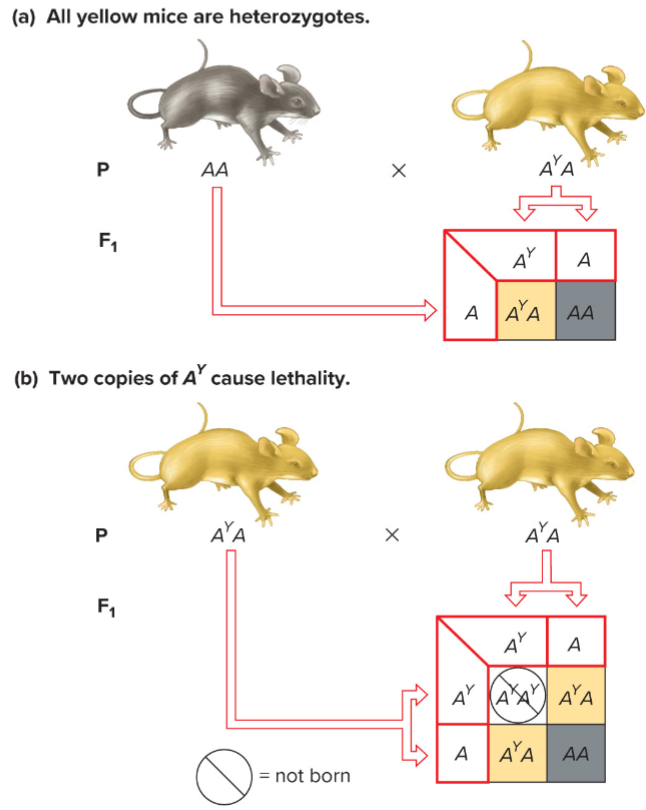
One gene, multiple phenotypes: Pleiotropy (lec 5)
unusual ratios w/ only 2 phenotypes
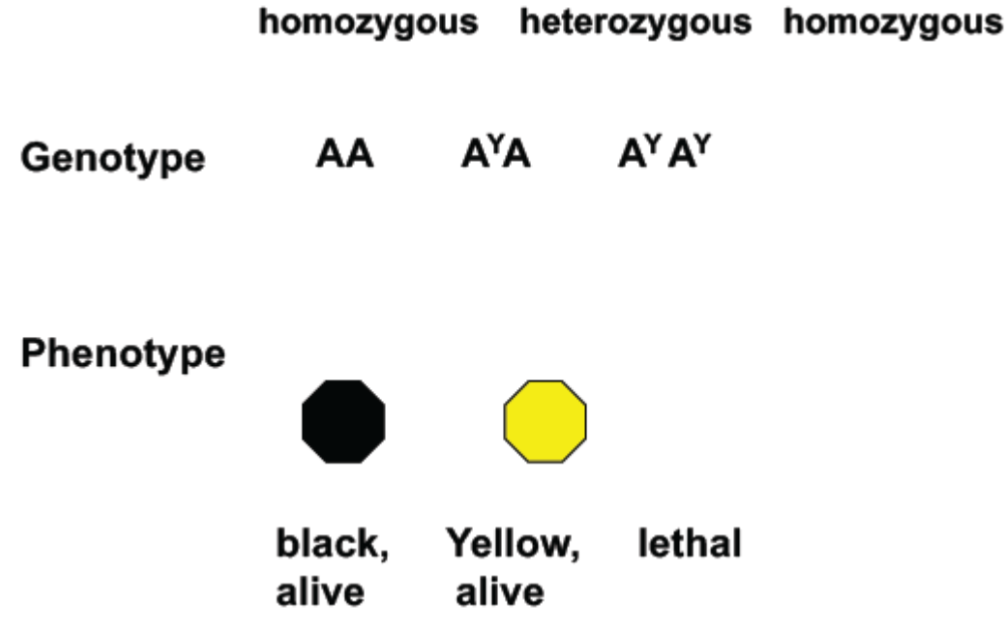
Understanding Pleiotropy: One gene controls multiple phenotypes (lec 5)
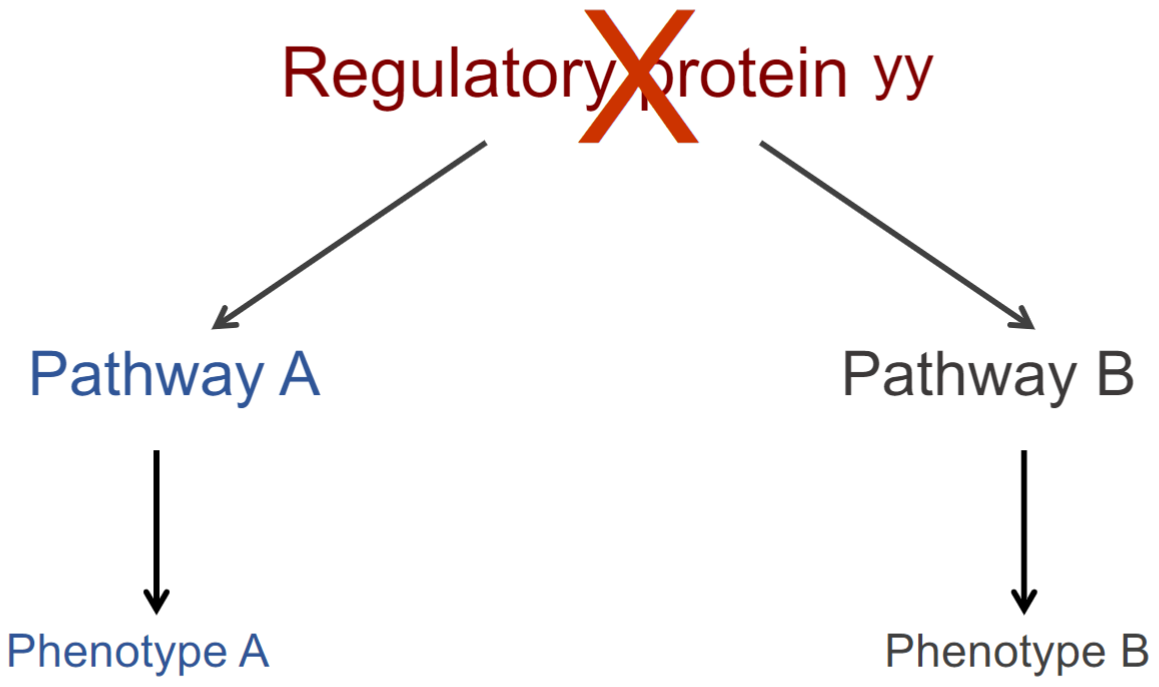
Pleiotropy (lec 5)
one gene has many symptoms or controls several functions
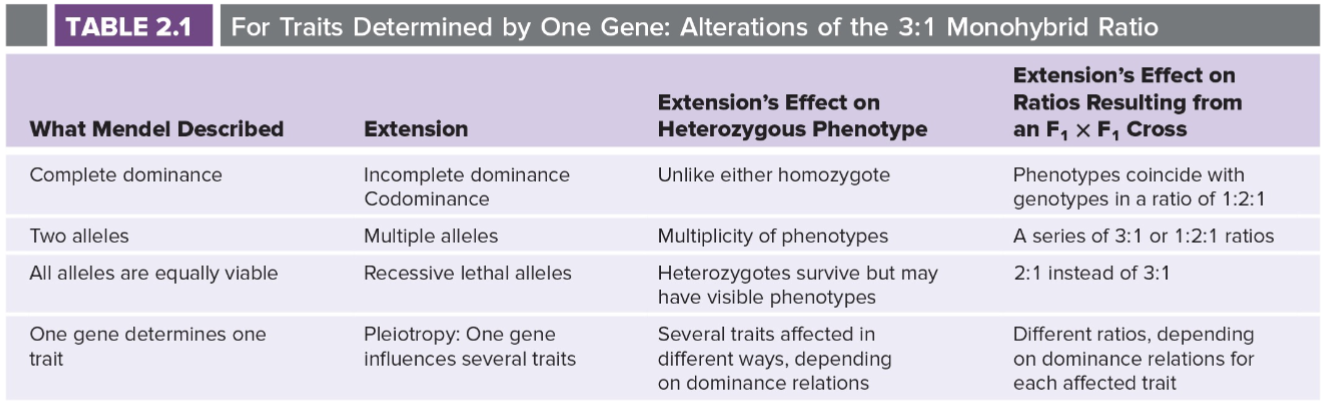
Extensions of Mendel’s laws: single genes (lec 5)
summary of Mendel’s basic assumptions and comparison of these assumptions w/ 20th century contributions
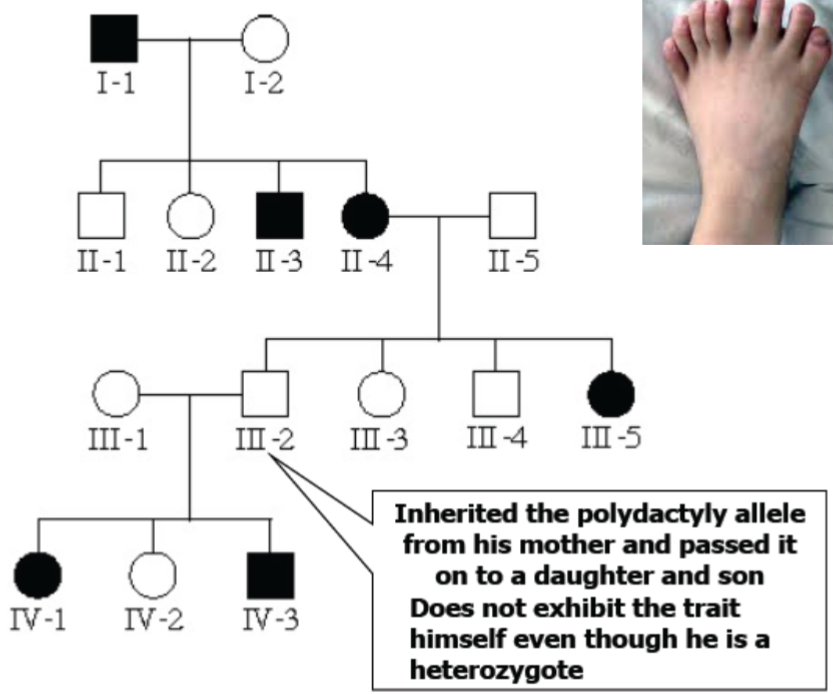
Other alterations to Mendel’s laws: Genotype isn’t always deterministic: Penetrance (lec 5)
For some genes, given genotype will give certain phenotype only some of the time (ex: 75% of homozygous recessive individuals will have disease)
individuals w/ same genotype can differ in phenotype due to incomplete penetrance
Penetrance:
is percentage of population w/ particular genotype that shows expected phenotype
can be complete (100%) or incomplete
Incomplete Penetrance example: polydactyly (lec 5)
Other alterations to Mendel’s laws: Genotype is not always deterministic: Expressivity (lec 5)
phenotypes can be expressed to diff degrees
ex: severity of disease can differ w/ same genotype
Expressivity: degree/intensity w/ which particular genotype is expressed in phenotype
can be variable/unvarying
Other alterations to Mendel’s laws: Phenotype isn’t always deterministic (lec 5)
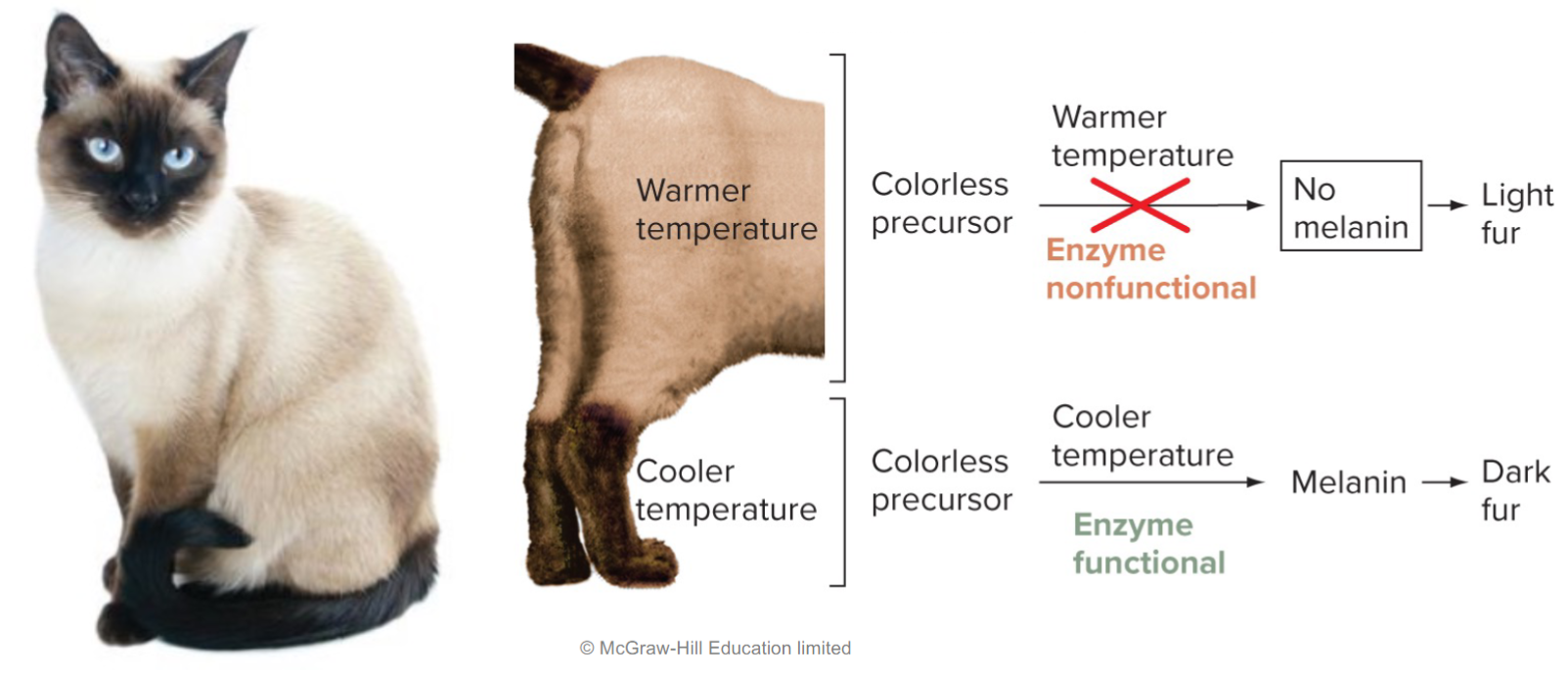
colour indicates level of expression (shades of green show varying expressivity)
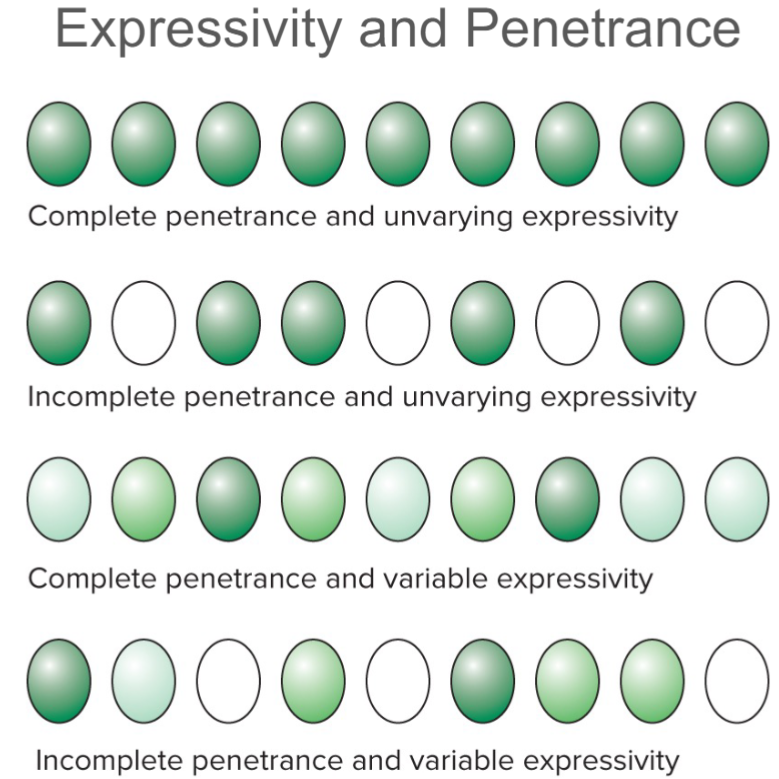
Other alterations to Mendel’s laws: Environmental modification (lec 5)
Genotype vs. Phenotype (lec 5)
Complicated relationships btwn genotype and phenotype can violate Mendel’s predicted 3:1 ratios
Mendel’s Laws of Segregation and independent assortment still hold
Extensions to Mendel for single-gene inheritance dominance isn’t always complete (lec 5)
crosses btwn true-breeding strains can produce hybrids w/ phenotypes diff from both parents
Incomplete dominance: ex: snapdragon flower colour
Codominance: ex: lentil coat patterns, ABO blood groups in humans, histocompatibility in humans
Pleiotropy:
recessive lethal alleles (ex: Ay allele in mice)
delayed lethality
Do variations on dominance relations negate Mendel’s Law of Segregation? (lec 5)
dominance relations affect phenotype and have no bearing on the segregation of alleles
alleles still segregate randomly during gamete formation
gene products control expression of phenotypes differently
Mendel’s Law of Segregation still applies
interpretation of phenotype/genotype relationship is more complex
Genetic interactions (lec 6)
more than just a few genes in genome
diff genes are likely to be interacting/dependent on each other to give rise to specific traits
can that explain deviation from Mendelian segregation?
Reciprocal Recessive Epistasis (lec 6)
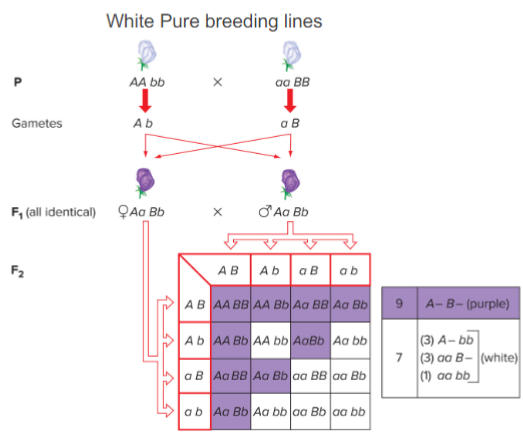
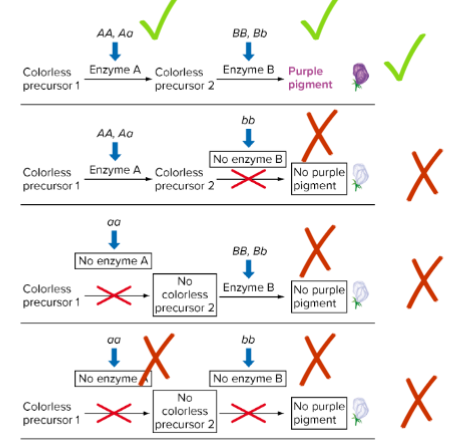
purple F1 progeny of sweet peas are produced by crossing 2 pure-breeding white lines
F2: 7 white:9 purple ratio
indicates 2 genes at work
purple: at least 1 dominant in both alleles (both enzymes must work)
White: 1 loci must be homo recessive
Possible biochem explanation for Reciprocal recessive Epistasis for flower colour in sweet peas (lec 6)
Locus heterogeneity in humans: Ocular-cutaneous albinism (lec 6)
heterogeneity: mutations in 2 diff genes but cause same phenotype
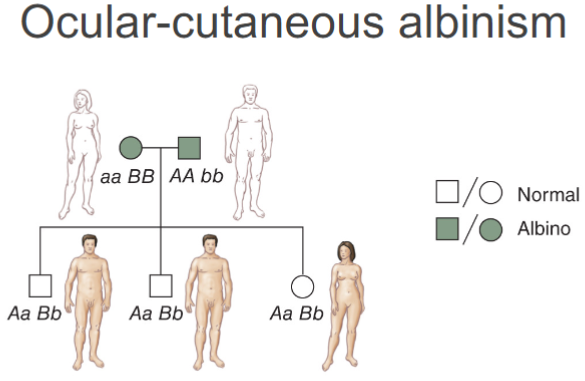
Inheritance of eye colour (lec 6)
blue recessive to brown
at least 12 genes affect eye colour, although 2 explain much of variation (H and O)
need at least 1 dominant allele for both genes for brown eyes (H-O-)
Locus heterogeneity in humans: complementation (lec 6)
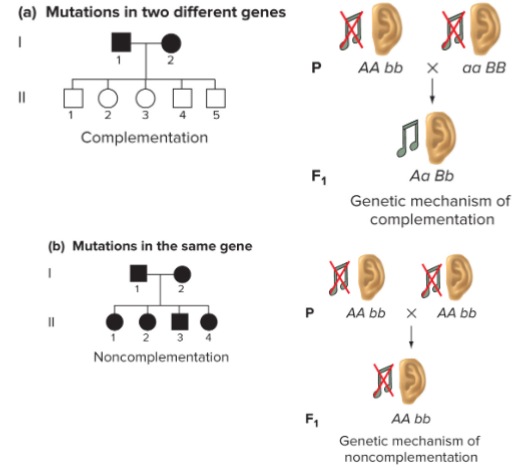
Complementation/Allelism (lec 6)
diff null mutations in same gene can abolish function
mutations in diff genes might also abolish function and give rise to same phenotype
how to distinguish btwn these possibilities?
Heterogeneous traits and complementation test (lec 6)
complementation testing used to determine if particular phenotype arises from mutations in same/separate genes
can be applied only w/ recessive, not dominant, phenotypes
“complementation group” synonymous w/ single gene
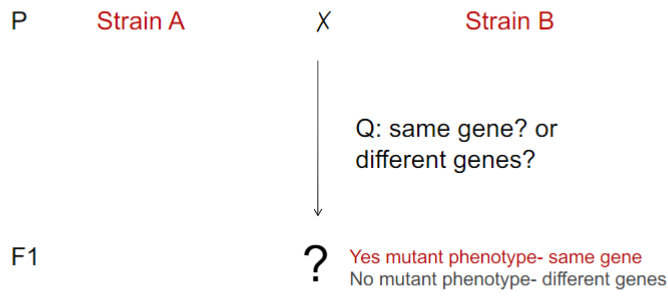
Complementation Test (lec 6)
works when phenotype is recessive
useful tool to determine how many genes contribute to trait (complementation groups)
mutant phenotype = same gene
no mutant phenotype = diff genes
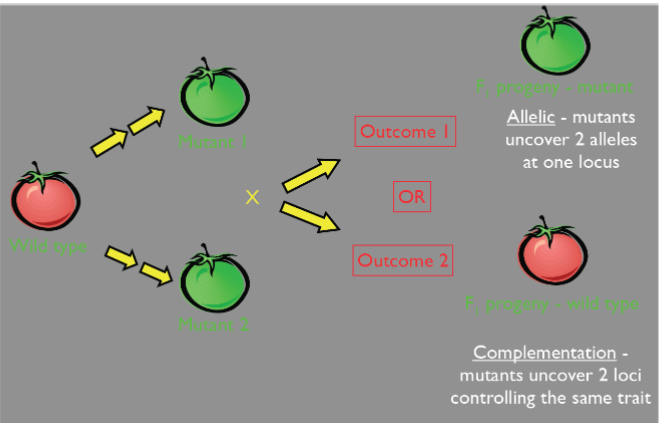
Many ways to “knockout” a gene: allelic variation (lec 6)
mutations can pop up in many places
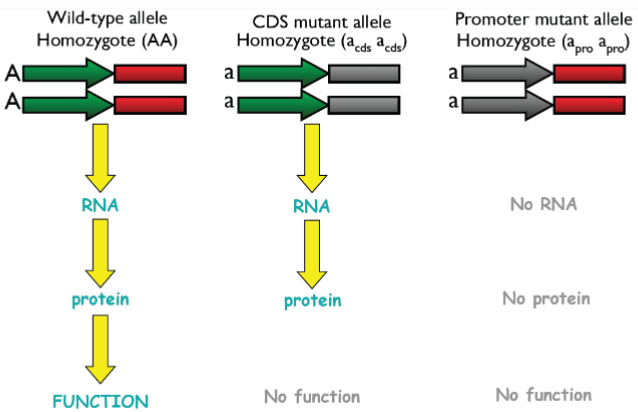
WT alleles complement mutant alleles (lec 6)
WT alleles complement in cases of haplosufficiency, phenotype is WT
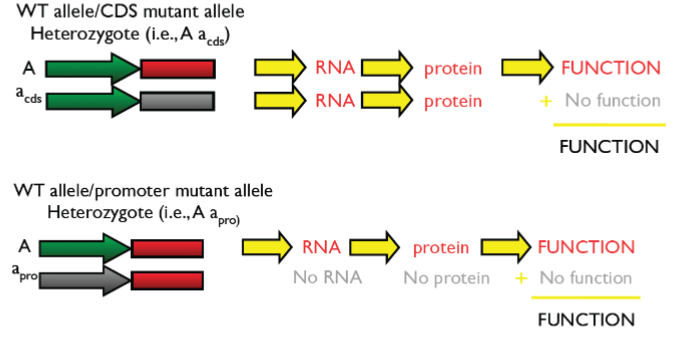
When mutant alleles at same locus can’t complement each other: they are allelic
2 mutant alleles can’t complement each other so phenotype is mutant (indicator of allelism)
Molecular vs Phenotypic visibility of allelism:
while locus appears hetero at molecular lvl, the individual appears homo at phenotypic lvl
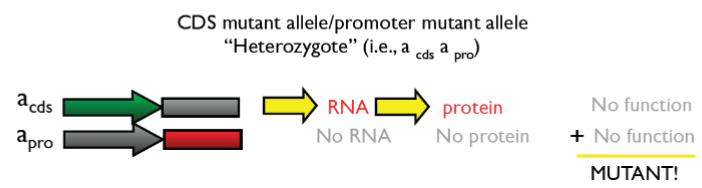
Forward genetics (lec 6)
identify mutants that produce target phenotype
complementation test: Cross inbred lines to test whether mutations are in same/diff genes
do genes interact?
Complementation test: sorting allelic from non-allelic (lec 6)
variety of molecular deficiencies, still allelic if in same gene
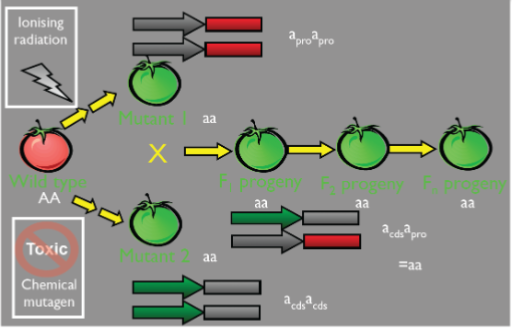
Complementation test: sorting allelic from non-allelic pt. 2 (lec 6)
2 loci = complementation