Biochem test 2
1/120
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
121 Terms
what are the three main functions of proteins?
structural - in cytoskeleton and collagen
enzymatic - bio-catalysts
binding - bind for transport, storage, or signaling
explain the lock and key model
the active site on an enzyme has a fixed, rigid geometrical shape. only a substrate with a matching shape can bind.
explain the induced fit model
the initial binding of a ligand induces conformational changes in protein to cause tighter binding.
how do enzymes function?
reversible binding of other molecules (ligands)
what is a ligand?
a molecule that binds only to the binding site on a protein
dissociation constant
the rate of dissociation between protein and ligand; high Kd indicates low ligand affinity; the concentration of ligand when half of the binding sites are bound
association constant
the rate of assembly between protein and ligand; high Ka indicates high ligand affinity
myoglobin
protein that contains 8 alpha helical segments; binds oxygen with heme prosthetic group
protoporphyrin
the planar complex organic ring found in several proteins such as myoglobin and hemoglobin; coordination of iron causes it to no longer be planar
where are the histidine residues in myoglobin?
His F8 (proximal) and His E7 (distal)
where does oxygen bind in myoglobin?
to the iron opposite of His F8
what is free heme?
heme outside of myoglobin that binds to CO and NO, which both have a higher affinity for Mb than O2 does
what is P50?
Kd on a pO2 x-axis where 50% of binding sites are bound
what subunits does hemoglobin contain?
2 alpha subunits and 2 beta subunits (a dimer of dimers)
explain the T state and R state of hemoglobin.
the T state (deoxyhemoglobin) dominates in the absence of oxygen. the R state dominates in the presence of oxygen. the T state is a low affinity state, and the R state is high affinity.
what is the hill equation?
used to determine cooperativity log(Y/1-Y) = nlog(L) - logKd
n = 1: no cooperativity
n>1: positive cooperativity
n<1: negative cooperativity
if n>1, n gives us the minimum number of binding sites on the protein.
explain the Bohr effect.
essentially, low pH stabilizes the T state, and vice versa because high pH’s deprotonate/destabilize the T state
immune response
a coordinated set of interactions among many classes of proteins, molecules, and cell types that distinguishes the molecular self from the nonself and destroys the nonself
leukocytes
white blood cells, including macrophages and lymphocytes
humoral immune system
1 of 2 complementary systems; directed at bacterial infections and extracellular viruses
cellular immune system
1 of 2 complementary systems; destroys infected host cells, parasites, and foreign tissues
immunoglobulins (Ig)
antibodies; bind bacteria, viruses, or large foreign molecules and target them for destruction; produced by B cells; 4 polypeptide chains; cleavage with protease papain releases the basal fragment; constant domains contain the immunoglobulin fold motif
cytotoxic T cells
recognize infected cells or parasites using T cell receptors
helper T cells
produce soluble signaling proteins called cytokines; interact with macrophages and stimulate the selective proliferation of cytotoxic T cells and B cells that can bind to a particular antigen
memory cells
permit a rapid response to pathogens previously encountered
antigen
molecule or pathogen capable of eliciting an immune response; antibodies bind to antigenic determinant or epitope in the antigen
haptens
small molecules that can elicit an immune response when covalently attached to large proteins
variable domain of Ig
associate to create the antigen-binding site; allows for formation of antigen-antibody complex
polyclonal antibodies
produced by injecting a protein into an animal; contain a mixture of antibodies that recognize different parts of the protein
monoclonal antibodies
synthesized by a population of identical B cells; all recognize the same epitope
explain the steps of ELISA.
coat the surface with the sample antigens.
block unoccupied sites with nonspecific proteins.
incubate with primary antibody against specific antigen.
incubate with secondary antibody-enzyme complex that binds primary antibody.
add substrate.
formation of colored product indicates presence of specific antigen.
primary motor proteins
actin and myosin; make up more than 80% of the protein mass of muscle. involved in ATP driven conformational changes that result in muscle contraction.
myosin
contains 2 heavy chains and 4 light chains; forms a fibrous, left-handed coiled coil domain on the tail and a large globular domain on the head
actin
associates to form a long polymer called F-actin
thick vs thin filaments
thick filaments: rodlike structures of aggregated myosin
thin filaments: F-actin along with proteins troponin and tropomyosin; assemble as successive monomeric actin molecules add to one end; each monomer hydrolyzes ATP
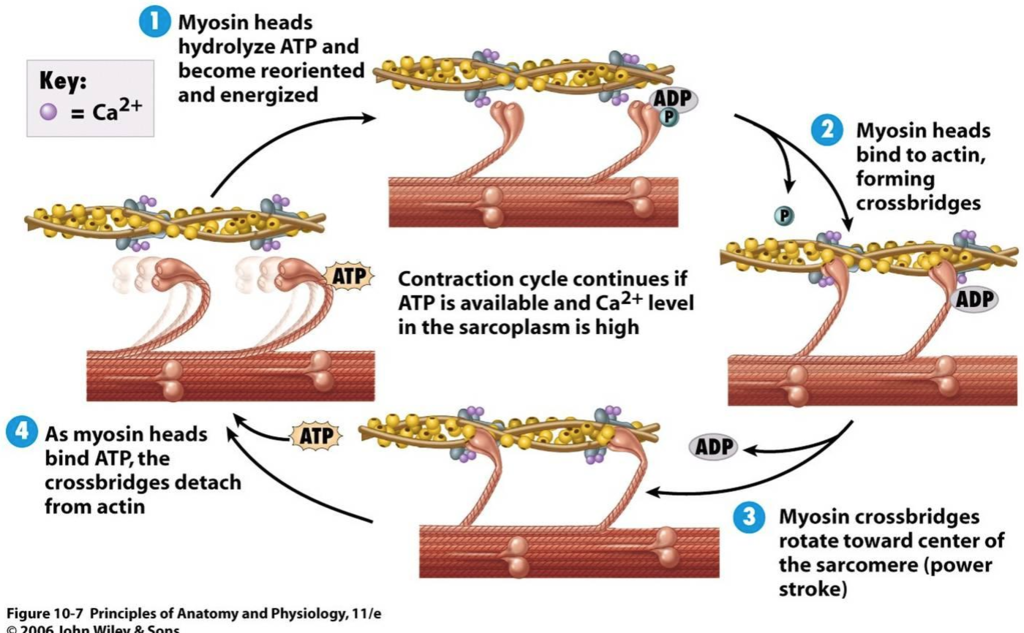
how do actin and myosin interact?
each actin monomer in the thin filament binds to one myosin head group. myosin binds to actin tightly when not bound to ATP. Undergo a series of conformational changes due to binding, hydrolysis, and release of ATP and ADP causing muscle contraction.
muscle fiber
large, single, elongate multinuclear cell; contains ~1000 myofibrils, each consisting of thick and thin filaments, and surrounded by sarcoplasmic reticulum
sarcomere
an entire contractile unit
A band
stretches the length of the thick filament
I band
contains only thin filaments
Z disk
attachment site for thin filaments
M line
bisects the A band
write out the steps of muscle contraction.
ATP binds to myosin head, causing dissociation from actin.
As tightly bound ATP is hydrolyzed, a conformational change occurs. ADP and Pi remain associated with myosin head.
Myosin head attached to actin filament, causing release of Pi.
Pi release triggers a power stroke. This is a conformational change in the myosin head that moves actin and myosin filaments relative to one another. ADP is released.
tropomyosin
binds to the thin filament and blocks the myosin binding sites
troponin
binds Ca2+ released from the sarcoplasmic reticulum, causes a conformational change, and exposes myosin-binding sites
ribozymes
RNA molecules that act as enzymes
cofactors
inorganic molecules like metal ions necessary for enzyme function
coenzymes
organic molecules necessary for enzyme function
holoenzyme vs. apoenzyme
holo: enzyme with cofactor
apo: enzyme with no cofactor
what do the 4 numbers in the enzyme classification system represent?
#1: enzyme class
#2: enzyme subclass
#3: enzyme group
#4: molecule/substrate
acid-base catalysis
1 of 3 catalysis mechanisms; involved BL acids and bases (proton transfer)
covalent catalysis
1 of 3 catalysis mechanisms; lewis acid base reactions; nucleophilic attack on an electrophile that leads to formation of a new bond
metal ion catalysis
1 of 3 catalysis mechanisms; metal ion stabilization of a charged transition state group or redox reactions
What is the relationship between Gibbs free energy and the k constant in enzyme kinetics?
As G increases, k decreases, and vice versa
What are the Michaelis-Menten assumptions?
The rate-limiting step is downstream from the initial step.
The substrate concentration is significantly larger than the enzyme concentration by ~1000x.
The enzyme-substrate complex concentration is constant.
Explain how initial velocity is determined.
An enzyme cocktail of the same volume is added to multiple test tubes. Contains enzymes, cofactors, and anything else necessary.
Prepare the substrate stock solution.
Add DI water to the test tube volume depending on how much stock solution is in each.
Monitor with UV-Vis
Plot velocity vs. substrate concentration.
The only thing changing is the amount of substrate in each test tube.
Vmax
theoretical maximum initial velocity of the system with units the same as initial velocity
Km
substrate concentration at half of Vmax
What is the Michaelis-Menten equation?
V0 = (Vmax * [S])/(Km + [S])
How are Km and Kd related?
Km gives a rough estimate of Kd of the E*S
What is the Lineweaver-Burk plot?
the double reciprocal of the MM equation
used to make a linear graph
How do you interpret a LB plot in terms of Vmax and Km?
lower 1/Vmax = higher Vmax (found at y-intercept)
lower -1/Km = lower Km ( found at x-intercept)
how do you find kcat?
Vmax/[E]
what is kcat?
the turnover #; defines the rate of the rate-limiting step
what is the specificity constant equation?
kcat/Km
what is special about regulatory enzymes?
they typically don’t follow Michaelis-Menten kinetics.
what are the 4 types of inhibitors?
competitive
noncompetitve
uncompetitive
mixed
how does a competitive inhibitor work?
it binds to the active site; doesn’t change Vmax; changes Km
how does an uncompetitive inhibitor work?
it binds to an allosteric site, but only after substrate is bound; changes Km and Vmax by the same order
how does a noncompetitive inhibitor work?
it binds to an allosteric site, either before or after substrate is bound; doesn’t change Km, does change Vmax
how does a mixed inhibitor work?
it binds to an allosteric site, either before or after substrate is bound; changes both Km and Vmax, but not by the same order
dead-end inhibitors
bind at the active site with no physical change; irreversible
suicide inhibitors
bind at the active site and mimic the natural substrate; begins catalysis but stops partially through, leaving something blocking the site; irreversible
transition state analogs
mimic the transition state of the substrate; bind at the active site very tightly; irreversible
feedback inhibitioin
noncovalent binding of a downstream product to the enzyme to upregulate or downregulate production
reversible covalent modification
covalent modification of a specific functional group
proteolytic cleavage
precursor proteins are acted upon by a specific protease which cleaves part of the peptide off of the precursor; active enzyme is left
what are the 5 regulators of glycolysis?
AMP, ADP, ATP, citrate, and fructose-2,6-bisphosphate
What are the 2 functional groups used to name monosaccharides?
aldehyde - aldose
ketone - ketose
what is the range of the number of carbons in a monosaccharide? what length is the most common?
3-7; 5-6
where are L-aldohexoses found?
peptidoglycan
where is 1 chiral carbon in monosaccharides always found?
the 2nd to last carbon; this carbon determines D or L configuration
what is a hemiacetal?
contain an R group, a hydrogen, an alcohol, and an ether group on the chiral carbon
what is a hemiketal?
contain 2 R groups, an alcohol, and an ether group on the chiral carbon
what are the difference in alpha and beta ring structures in carbohydrates?
alpha structures contain the anomeric -OH below the ring; beta structures contain the anomeric -OH above the ring
pyran
sugars that form a 6-membered ring
furan
sugars that form a 5-membered ring
what is another word for polysaccharides?
glycans
starches
consist of amylose and amylopectin; in plants; homopolymers of glucose
glycogen
amylopectin with more branches; homopolymer of glucose; in human liver
cellulose
homopolymer of glucose; structural support for woody plants; beta 1-4 linkages for stability; has stabilizing H-bonds to leave no room for water
chitin
homopolymer of N-acetylglucosamine; main component of exoskeleton of arthropods; identical to cellulose except contains acetylated amino groups on C2 carbon
proteoglycans
macromolecules of cell surface containing 1+ glycosaminoglycan chains that are covalently bound to a membrane/protein
what is the enzyme for the reaction: Glucose + ATP —> ADP + Glucose-6-phosphate?
hexokinase
What is the enzyme for the reaction: Glucose-6-phosphate —> Fructose-6-phosphate?
phosphatase isomerase
what is the enzyme for the reaction: Fructose 6-phosphate + ATP —> ADP + Fructose 1,6-bisphosphate?
phospho-fructokinase-1
What is the enzyme for the reaction: Fructose 1,6-bisphosphate —> glyceraldehyde 3-phosphate + dihydroxyacetone phosphate?
aldolase
what is the enzyme for the conversion of dihydroxyacetone phosphate to glyceraldehyde 3-phosphate?
triose phosphate isomerase
What is the enzyme for the reaction: 2 glyceraldehyde 3-phosphate + 2 Pi + 2 NAD+ —>2 NADH + 1,3-bisphosphoglycerate (2)?
glyceraldehyde 3-phosphate dehydrogenase
what is the enzyme for the reaction: 1,3-bisphosphoglycerate (2) + 2 ADP —> 2 ATP + 3-phosphoglycerate (2)
phosphoglycerate kinase