CRISPR genotyping
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
23 Terms
Multiplex PCR
Standard polymerase chain reaction (PCR) amplifies a specific nucleotide sequence (amplicon) using a single set of primers (usually a pair). Multiplex PCR is the simultaneous amplification of multiple amplicons in a single reaction using a unique primer set for each (Figure 1). As with standard PCR, a primer set targets a particular DNA sequence. Either the absence of a target PCR sequence or the disruption of primer binding sites will prevent amplification of the target PCR sequence. If an amplicon is detected in a sample, often by agarose gel electrophoresis, then the associated target DNA sequence was present in the sample
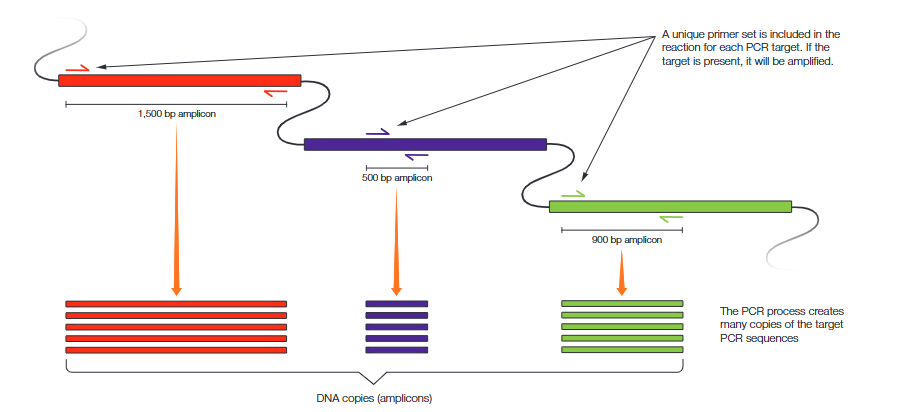
Using Multiplex PCR to Detect Gene Editing
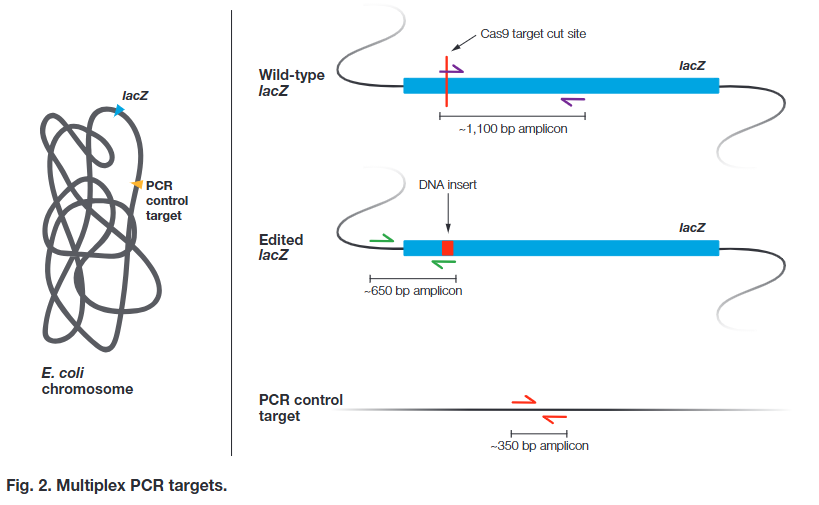
Multiplex PCR can be used to confirm these conclusions at the DNA level. After a quick DNA extraction from each colony, you will use multiplex PCR to detect the presence of the donor template DNA insertion. The multiplex PCR reaction includes three primer sets (Figure 2):
• The first primer set is designed to detect unmodified lacZ. One primer in the set will bind directly to the Cas9 target cut site. If the target cut site was modified, the primer will not bind. If the target Cas9 cut site was NOT modified, then this primer set will yield a ~1,100 bp amplicon
• The second primer set is designed to detect modified lacZ. One primer in the set will bind to the insert from the donor template DNA. If the target cut site was successfully repaired using the donor template DNA that was introduced to the bacteria, then this primer set will yield a ~650 bp amplicon
• A third set of primers will amplify an unrelated region far downstream of the lacZ gene as a control to verify that chromosomal DNA is present in the sample. If chromosomal DNA was successfully extracted and PCR was successful, whether or not lacZ was modified, then this primer set will yield a ~350 bp amplicon
State the results of the controls in your experiment and describe what you can conclude from them
The results of the control amplifications provide assurance that the DNA extraction and PCR processes were successful and validate the presence of chromosomal DNA in the samples. If the control amplifications yield the expected amplicons, it confirms the reliability of the multiplex PCR results related to the gene editing.
Do the results from the multiplex PCR disprove either of your claims (your original claim or your alternative claim) from Part 1? Explain why or why not
No, my clain was that CRISPR widiting would result in a functional lacZ gene however this was not disproves since our D3 results were not what was expected
Write a new claim or set of claims, based on evidence, about the role of CRISPR gene editing in modifying the lacZ gene in bacteria grown on experimental plates C and D. Include evidence from both your bacterial transformation and multiplex PCR experiments
The new claim is that CRISPR gene editing effectively modifies the lacZ gene in bacteria, leading to differential expression observed in experimental plates C and D. Evidence from the bacterial transformation shows successful incorporation of donor template DNA, while multiplex PCR confirmed the presence of modified lacZ, indicating that the targeted gene editing was successful.
Amplicon
a piece of DNA that was produced through amplification, often by PCR. A PCR product can be called an amplicon
Base pair (bp)
complementary nucleotides held together by hydrogen bonds
Cas9
CRISPR-associated protein 9, an endonuclease that forms a double-strand break (cut) in DNA at a specific site within a larger recognition sequence, or target site. Involved in the natural defense of certain prokaryotes against DNA viruses, it is also heavily utilized in genetic engineering applications to cut DNA at locations specified by a guide RNA (gRNA)
CRISPR
clustered regularly interspaced palindromic repeats are sequences in the genomes of some prokaryotes that act as a genomic record of previous viral attacks. Along with CRISPR associated (Cas) proteins, bacteria use the sequences to recognize and disarm future invading viruses. Scientists have adapted this system for genetic engineering purposes
Chromosome
— a DNA molecule with all or part of the genetic material for an organism.
Donor template DNA
a sequence of DNA required for homology-directed repair in CRISPR gene editing applications; may include a desired sequence flanked on both sides by “homology arms” that match the sequence upstream and downstream of the cut
Gene editing
manipulation of genetic material in living cells by adding, removing, and replacing DNA sequences, typically with the aim of altering phenotypes.
InstaGene Matrix
microscopic beads that bind divalent cations in solution; the binding or sequestering of divalent cations prevents their availability to enzymes that can degrade the DNA template
lacZ
part of the lac operon in E. coli, this gene encodes the enzyme β-galactosidase. For decades, molecular biologists have used the lacZ gene as a target site for inserting DNA sequences because the bacterial colony color indicates whether any edits were successful
Master mix
the main reagent solution used in PCR, master mix contains all the necessary components (dNTPs, primers, buffer, salts, polymerase, polymerase cofactor)
Multiplex PCR
the simultaneous amplification of multiple targets by polymerase chain reaction. Multiple primer sets are used in the reaction, one for each target.
Polymerase chain reaction (PCR)
the process of amplifying or synthesizing DNA in vitro using primers and cycles of temperature change.
Primer
a short sequence of nucleotides (usually 16–24 bases in length) that recognizes a particular sequence of nucleotides on the target DNA sequence; primers for the polymerase chain reaction are usually synthesized in a laboratory.
X-gal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, a compound consisting of galactose linked to a substituted indole. Its hydrolysis by β-galactosidase yields an insoluble blue pigment that indicates the presence of active β-galactosidase.
What is genotyping
Genotyping means figuring out what version of a gene someone has—like reading a specific chapter in a book to see how it’s written
What are you doing in the lab?
You take a sample (usually cells), extract DNA, and then use a method called PCR (Polymerase Chain Reaction) to copy a specific part of the DNA millions of times.
Then you use gel electrophoresis to visualize differences in DNA length or pattern between samples.
In this experiment, you’re testing to see if a gene involved in fungal growth has been successfully edited using CRISPR.
Why do this?
To confirm whether CRISPR editing worked by comparing edited and unedited DNA.
What does it teach?
How to extract DNA
How PCR amplifies specific genes
How gel electrophoresis separates DNA fragments
How scientists check if gene editing or mutations occurred