03 - Genome Maintenance and Change
1/99
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
100 Terms
do closely related eukaryotes have simmilar genome sizes
some closely related eukaryotes have large differences in genome size
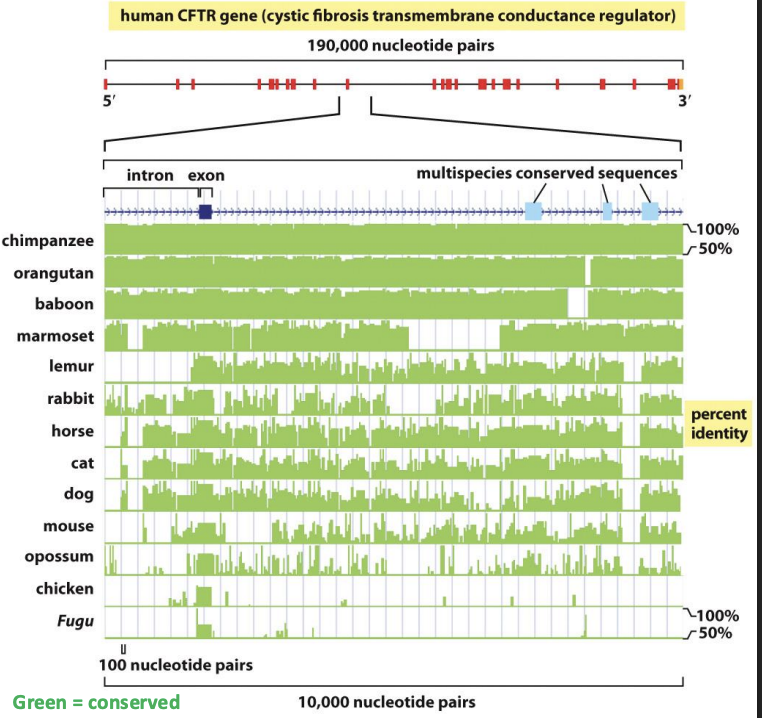
What does this cross-species comparison of a gene sequence tell us about the accumulation of genetic change?
even within a gene, some sequences are more highly conserved
exons are conserved
Do all sequences accumulate change at the same rate?
no
in very important genes = mutations very harmful = dont help us and are removed = so they wouldnt persist = limmited change
most imp genes = highly conserved
what does highly conserved genes mean
highly important
Would all sequences encoding amino acids in a protein be equally important/conserved?
they are not conserved equally, some sequences are more important than others
eg active sites if you chnag eit will destroy
Multispecies sequence comparison show:
Protein-coding sequences of genes are more highly conserved than genome size and organization (including # of basepairs, # of chromosomes, # of genes, position of genes along chromosomes, amount of repetitive DNA)
Exons are more highly conserved than introns
Exons
(coding for the amino acid sequence of proteins)
introns
(spliced out during RNA processing)
Mutation
change in the DNA sequence
what can cause mutation
errors in DNA replication or repair
point mutation
switching one nucleotide for another
which error rate is higher bacteria or human
bacteria
Large-scale rearrangements in change in dna sequence
Insertions Deletions Duplications Inversions Translocations
what mechanism of change is likely to result in loss/mutation of genes?
Deletions
intrachromosomal rearrangement
within 1 chromosome
interchromosomal rearrangement
between 2 chromosomes
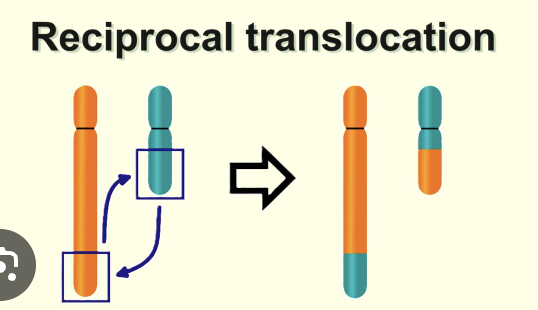
reciprocal translocation
swapped without any net loss or gain to the genome
reciprocal exchange
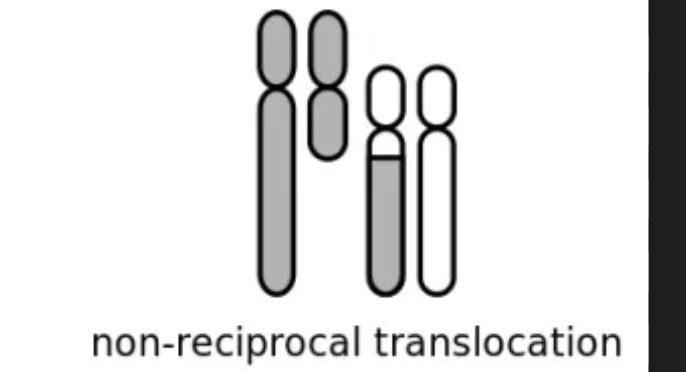
non reciprocal translocation
one-way translocations in which a chromosomal segment is transferred to a nonhomologous chromosome
no reciprocal exchange
synteny
These blocks that carry genes in conserved order (but now located in different positions)
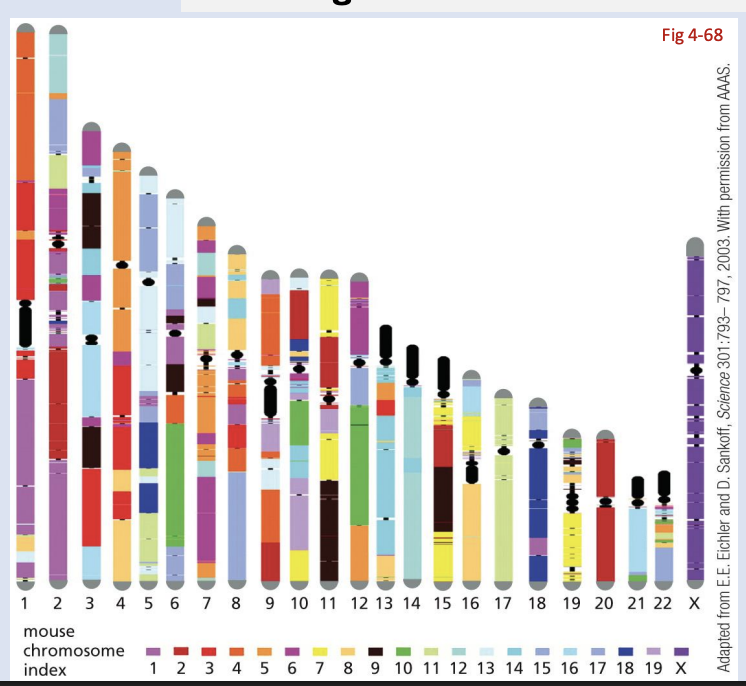
what do you think is more highly conserved: sequences within exons or the positions of genes on particular chromosomes?
exons sequences are more conserved
positions move around and scrambled
what percent of human genome shows conservation across species.
5%
Gene sequences only make up about a third of that; There are many conserved DNA sequences of unknown function
Highly Repetitive DNA
At least 105 copies of the sequence and make up 1-10% of a genome
Tandem repeats
repeat over and over without interruption
Satellite DNA
(generally found at centromeres and telomeres)
- 5-500 bp in tandem repeats of up to 100 kb that can form very large clusters.
Base composition is distinct from bulk DNA
Minisatellite DNA
10-100 bp with up to 3000 repeats → highly variable (polymorphic), therefore # repeats differs between individuals– basis for DNA fingerprinting in criminal and paternity tests
Microsatellite DNA
1-5 bp in clusters of 10-40 bp scattered quite evenly throughout the genome → highly variable (# of repeats changes fast!), can be used to compare closely related populations
Moderately Repetitive DNA
• 20-80% of the genome depending on the organism
• Repeated a few times to tens of thousands of times
• Can include genes or non-coding DNA
Moderately Repetitive - Genes
many copies of ribosomal RNAs and histones genes
Moderately Repetitive - Non-coding
Sequences scattered throughout the genome (interspersed) caused by transposons
SINEs
short interspersed elements
LINEs
long interspersed elements
slippage
In both prokaryotes and eukaryotes, repetitive sequences can be expanded or destroyed by “slippage”
events causing misalignment during replication
“Microsatellite instability”
related to progression of some diseases
Mobile DNA
DNA that moves from one place to another in the genome
genetic rearrangement is called transposition
moving mobile genetic elements were called transposable elements or transposons
DNA transposons move by
“cut-and-paste”
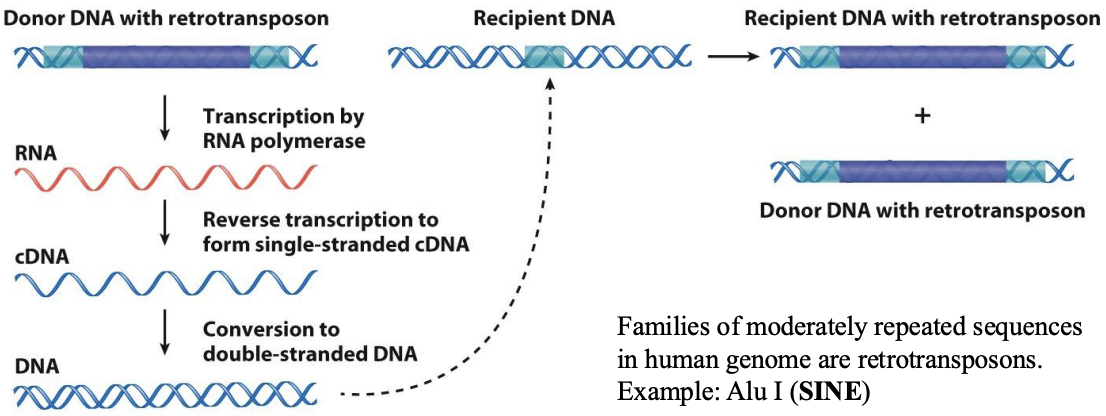
“Copy-and-paste”
what is “cut-and-paste” catalyzed by
catalyzed by the transposase enzyme
what is “cut-and-paste” mechanism
1. Inverted repeats on the ends of the transposon are required for recognition by transposase, which catalyzes excision from the donor DNA site and insertion elsewhere.
2. A direct repeat is generated in the recipient DNA site
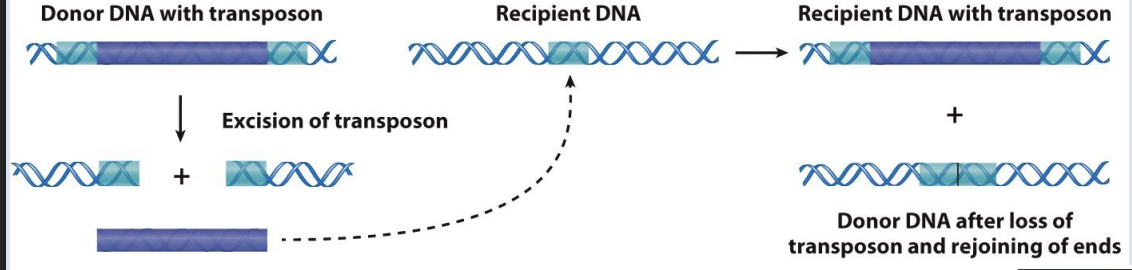
what is “Copy-and-paste” catalyzed by
RNA intermediate
what is “Copy-and-paste” mechanism
Retrotransposons use and can encode a reverse transcriptase enzyme to catalyze production of DNA from RNA. Example: LINEs and SINEs
Why do you think plants and animals can survive a higher frequency of transposition events?
survive high frequency bc of large genome with non-coding regions
transposons insert without disruption and minimize harmful effect
transposition in viruses
Some viruses use transposition mechanisms to move into host genomes and can also be a source of genetic change
Over time/generations, can different types of DNA sequences accumulate change at different rates
Can be because some sequences (ex. protein-coding) do not tolerate change without impacting health of the organism
Can be because some sequences are more prone to mutation (ex. misalignment in repetitive DNA)
exon shuffling
Beyond just mutation or disruption, transposition can also result in moving/adding or removing gene coding or regulatory sequences
how can new Genes Generated from Pre-existing Genes
Small substitutions, insertions, deletions can create different variants of genes
Gene duplication may can give rise to multigene families
Translocations can also create new genes (by exon shuffling mixing together different protein-coding segments)
Horizontal gene transfer (from other organisms/viruses) is another source of new genes
what results in gene duplication
Gene duplication can result from unequal crossing over and large chromosomal rearrangements
What happens when a change occurs that is very damaging to the organism?
dies an ddamages chain
What happens to changes in DNA of somatic cells?
not inherited by offsprings
orthologs
speciation gives rises to two separate species
paralogs
gene duplication and divergence
Gene “homologs”
(meaning sharing similarity because of common ancestry/origin
include orthologs and paralogs
Evolution of Globin gene
Globin gene family includes hemoglobin, myoglobin, and plant leghemoglobin
Pathway of globin gene evolution: Creation of a multigene family from a single ancestral gene via gene duplication, mutations, deletion and transposition
Pseudogenes
have a similar sequence to the gene family but have accumulated so many mutations that they are now non-functional.
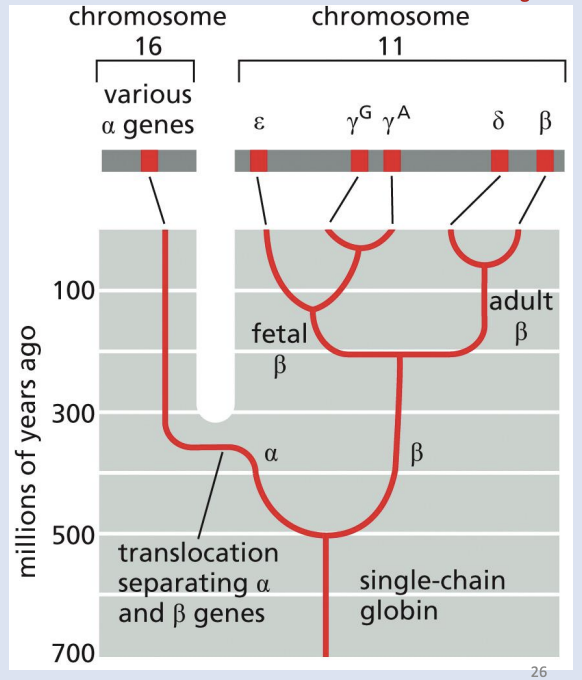
Which of the following are true about globin genes in humans?
A)They are paralogs
B)They are orthologs
C)They are homologs
D)They are alleles of each other
E) They are all pseudogenes
F) Comparing their DNA sequences will show a lot of similarities but also some divergence
a,c, f
Why do we care about small genetic changes? Do they really matter?
• Human and chimpanzee genomes differ by ~4% • Some minor genetic differences can result in major phenotypic differences!
Genetic variations within human populations
Single Nucleotide Polymorphisms (SNPs, “snips”)
Copy number polymorphisms (CNPs)
Structural Variation
Single Nucleotide Polymorphisms (SNPs, “snips”)
• Most common variability among humans
• SNPs in protein-coding regions contribute to the phenotypic differences in humans (alleles, some linked to diseases, drug interactions, etc.)
Copy number polymorphisms (CNPs)
• Differences in the number of copies of a particular sequence
• In protein coding region, extra copies = more protein → phenotypic differences
Structural Variation
Large segments of the DNA can be changed by duplication, inversion, deletion etc.
Which of the following are TRUE about comparing DNA sequence of genomes across species (select all that apply):A) All regions of DNA accumulate change at the same rate
B) Exons are more highly conserved then introns
C) All conserved sequences encode for proteins
D) Protein-coding sequences are more highly conserved than the location of genes on particular chromosomes
E) Highly repetitive DNA is more highly conserved as it is easier to replicate without errors
b, d
You want to use comparisons of genomic DNA for the purposes of identification or phylogenetic classification. Select the best strategy below:
A) Use the number of copies at a site of microsatellite DNA to trace species divergence from
the last common ancestor of bacteria and humans
B) Figure out if your poodle mix dog is more Bernedoodle, Bordoodle, or Springerdoodle by comparing the protein coding sequence of DNA polymerase
C) Use the number of repeats in minisatellite DNA to figure out who the father of an individual is (a paternity test)
D) Use the number of chromosomes in a blood sample to identify a murdered in a criminal investigation
c
Imagine 10 years from now you are working in medicine and you read a patient’s chart and tell them they will be switching to a different medication, because they have a SNP associated with a particular drug interaction. They say… “Huh?” and start asking you a LOT of questions and want to have a full lesson in genetics and molecular biology. Which of the following are TRUE things that could come up in this discussion? (select all that apply)
A) SNPs only occur in somatic cells, so don’t worry; they won’t be inherited by your children
B) SNPs can create different versions of a gene (also called alleles) that result in different traits in a person
C) You inherit one copy of each gene from each of your parents, but these might be different versions (alleles)
D) A SNP is a particular change or difference at 1 base pair in the sequence of your DNA
E) Proteins encode RNA which encode DNA, so damage to a protein results in a mutation in the associated gene in DNA
F) SNP stands for Separate Nuclear Polymorphism
b, c, d
What would happen to life on earth if DNA replication (and repair) were always perfect?
no variation
Perfect replication = no evolution = eventual extinction
What would happen to life on earth if DNA replication (and repair) were much more error-prone?
Too many harmful mutations → short-term survival risk
proposed models of dna replication
semi conservative replication
conservative replication
So which is it? Conservative, semi-conservative or dispersive? 34
DNA Replication is Semi - Conservative
DNA strand is synthesized in what direction
5’ to 3’
Incoming nucleoside triphosphate base pairs with the _____ strand and is then covalently attached to the ___
template
3’ OH
what provides the energy to create this new bond
hydrolysis of the bond between phosphates
Could NOT work if trying to extend the 5’ phosphate)
Replicating DNA with a low error rate - how
DNA polymerases add new nucleotides to a growing DNA strand in the 5’ to 3’ direction
1 existing DNA strand serves as a template, and if an incorrect nucleotide addition disrupts complimentary base-pairing, then it is usually detected and corrected
Proofreading function of DNA polymerase:
• DNA polymerase has a separate catalytic sites for polymerization and for editing
• DNA is pushed into the editing site when a mismatch is detected
three Proofreading mechanisms during replication
5’ → 3’ polymerization
3’ → 5’ exonucleolytic proofreading
strand- directed mismatch repair
Strand directed mismatch repair (MMR)
1. Recognition of mismatch by MutS
2. Identification of newly synthesized strand
3. Removal of incorrect nucleotides from new strand
4. Resynthesis of excised segment (using other strand as template)
5. Ligation to seal DNA backbone
what is used for recognition of mismatched base-pairing
Distortions of the double-helix
Why edit the newly synthesized strand instead of the template strand?
Ability to reversibly make single-stranded DNA (and use it as a template) is critical for transcription and DNA replication AND allows us to probe and synthesize DNA in the lab
How does the structure of DNA make replication possible?
- Hydrogen bonds between bases allow DNA strands to be separated.
- Opening up DNA (making it single stranded) allows each parent strand to serve as template for synthesis of a new complimentary strand.
- Complimentary base-pairing allows the selection of the correct nucleotides to properly replicate the sequence.
Renaturation also referred to as
Reannealing or Hybridization
process of DNA Denaturation and Renaturation
dna double helices → heated → denaturaturation to single strands (hydrogen bonds between nucelotide pairs broken) → slowly cool → renaturation restores dna double helices (nucleotide pairs reformed)
DNA Denaturation
Experimentally when DNA is slowly warmed to a certain temperature, DNA strand separation begins
Different DNA sequences denature at different temperatures
Thermal denaturation of DNA
DNA Melting
why is uv radiation useful for determining dna concentration
The nitrogenous bases of a nucleic acid absorb UV radiation (maximum at 260 nm), which can be used to determine DNA concentration
Single stranded (ss) DNA absorbs ___ as much UV light compared to double stranded (ds) DNA
twice
Melting Curve - y axis and x axis
UV absorbance on Y-axis, temperature on X-axis
Tm
the temperature at which the shift in absorbance is half completed
do all dna sequence denature at same temp
no
The higher the GC content of DNA, the ____ the Tm
higher
DNA Renaturation
Occurs when denatured DNA is allowed to cool and complementary sequences ‘find’ each other and base pairing begins
After renaturation, DNA is once again a double-stranded helical molecule
Renaturation Rates in diff size genome
Small genome = faster renaturation
Large genome = slower renaturation
how to make and measure a C0t plot
Shear DNA into equal-sized fragments – to avoid size effects on renaturation speed.
Heat the DNA to denature it (separate strands).
Cool slowly, allowing complementary strands to renature (hybridize).
Measure how much DNA has renatured over time.
C0 t value:
Combination of two values: 1. DNA concentration 2. Incubation time
• X-axis: C0 t plot
Initial DNA conc. (C0 ) X time of reaction (t)
• Y axis: C0 t plot
Fraction (percentage) of original DNA conc. that is renatured (dsDNA)
C0 t plot Curve shape:
* Single symmetrical curves because all sequences are present at the same concentration
Will repetitive sequences anneal faster or more slowly?
more faster
Experiment involves recording data on denaturation of DNA
“DNA Melting Curve” or “C0 t plot”
neither
Experiment involves recording data on DNA polymerization
“DNA Melting Curve” or “C0 t plot”
C0 t plot
Experiment involves recording data on DNA hybridization
“DNA Melting Curve” or “C0 t plot”
DNA Melting Curve
Experiment involves treating sample with various temperatures
“DNA Melting Curve” or “C0 t plot”
DNA Melting Curve
Experiment involves quantifying changes in the sample based on absorbance of UV light
“DNA Melting Curve” or “C0 t plot”
DNA Melting Curve
Results reflect the %GC base pairs in the DNA
“DNA Melting Curve” or “C0 t plot”
DNA Melting Curve
The shape of the results graph depends on whether or not the DNA contains a mix of both unique & repetitive sequences
“DNA Melting Curve” or “C0 t plot”
C0 t plot