Transcription - Lecture 7
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
68 Terms
Transcription (i.e. RNA synthesis)
required for gene expression
makes genes accessible
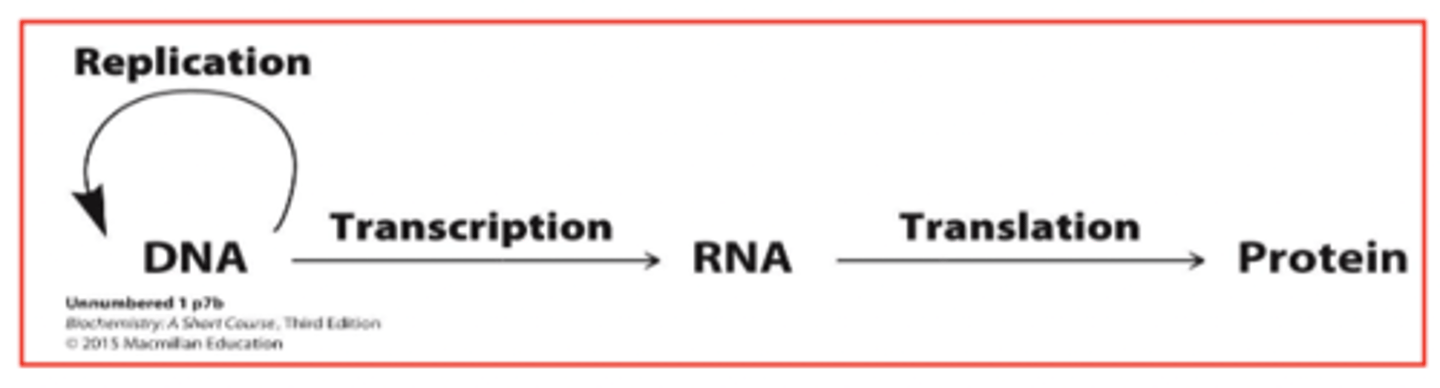
the central dogma
central information pathway in a cell
DNA makes RNA, RNA makes proteins
Where transcription occurs
occurs on a per-gene (or small group of genes) basis
Transcription - RNA synthesis
this does not require DNA synthesis to happen beforehand
Translation (i.e. protein synthesis)
transfers genetic information to proteins
DNA sequences
serve as physical and functional units of inheritance
DNA sequences - genes
people have two copies of most _____, but this number can vary
DNA sequences - variation
major source of _________ between individual genomes
DNA in somatic cells
in multicellular organisms, DNA is identical in all of these cells, but gene expression differs within the +200 different cell types
Gene expression
connects genotype (DNA) to phenotype (appearance)
Disease
often connected to changes in default gene expression level
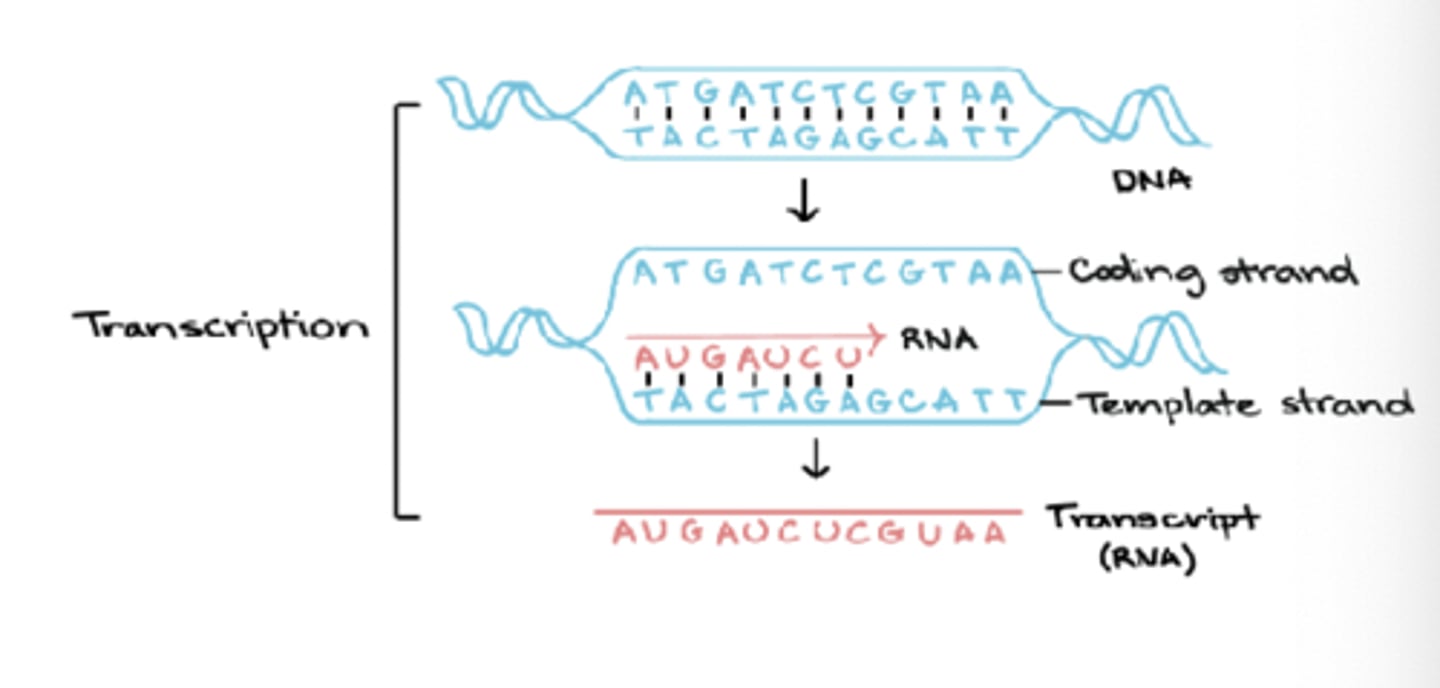
Synthesis of RNA (transcription) - direction
unidirectional as only one DNA strand used as a template
Synthesis of RNA (transcription) - location
occurs in cytoplasm (prokaryotes) or nucleus (eukaryotes)
Synthesis of RNA (transcription) - transcription start sites
this is at a specific DNA location
Synthesis of RNA (transcription) - "transcription unit"
each of these encodes at least 1 gene
Transcription stages
initiation, elongation, and termination
Denaturation
transcription requires DNA to undergo this
strands must be separated for cellular machinery to access nucleic acid
Denaturation location
occurs at a local region; entire helix does not unwind
B form to A form
DNA makes this transition as this conformation improves fidelity via template nascent and protein-DNA interactions
prokaryotes - RNA polymerase
in prokaryotes ___ __________ has helicase activity
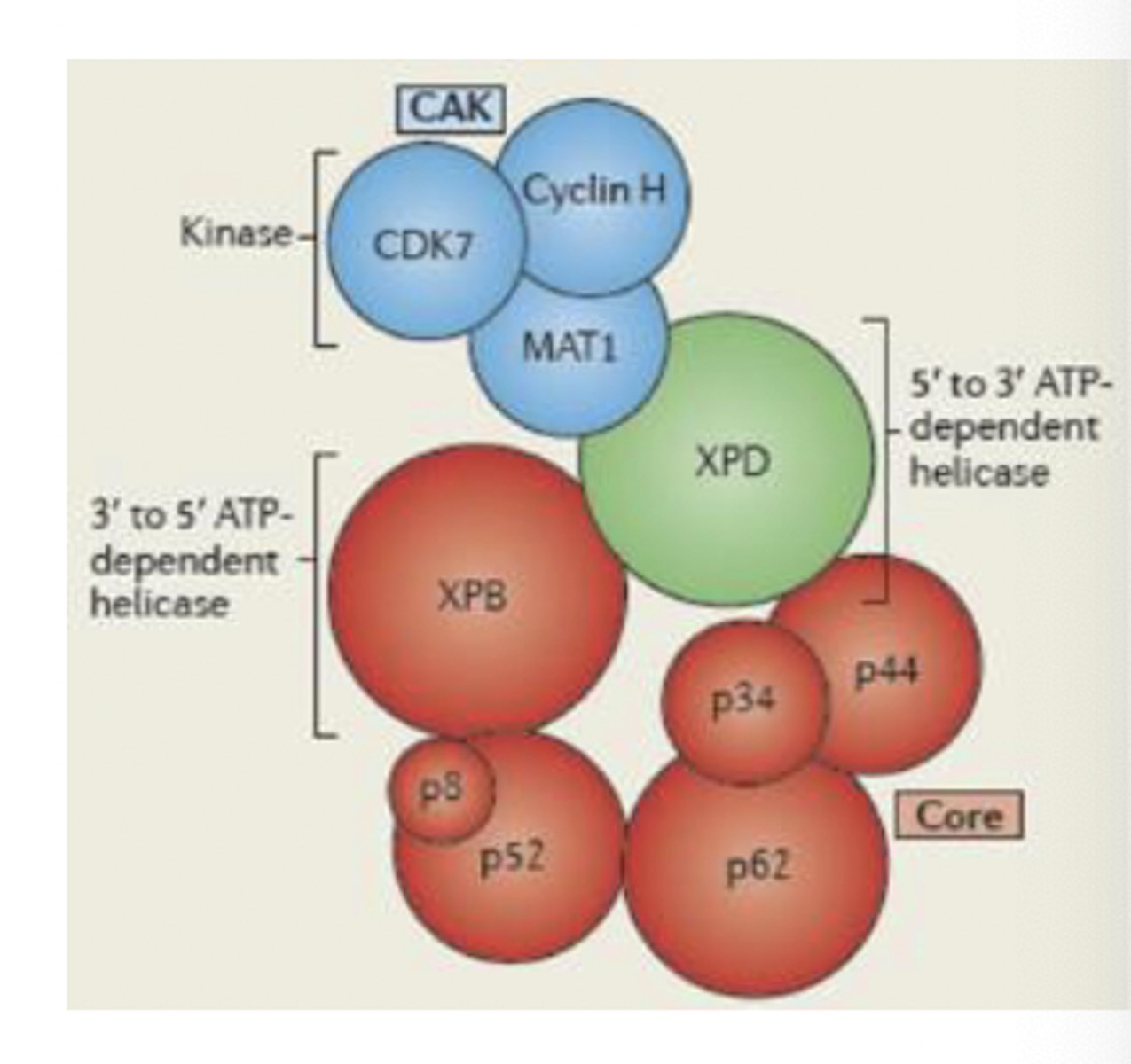
transcription factor IIH
in eukaryotes, this functions as a helicase for RNA polymerase II
RNA polymerase
key DNA-binding protein that catalyzes phosphodiester bridge formation in RNA
RNA polymerase - ribonucleotides
added to the 3' end of newly synthesized
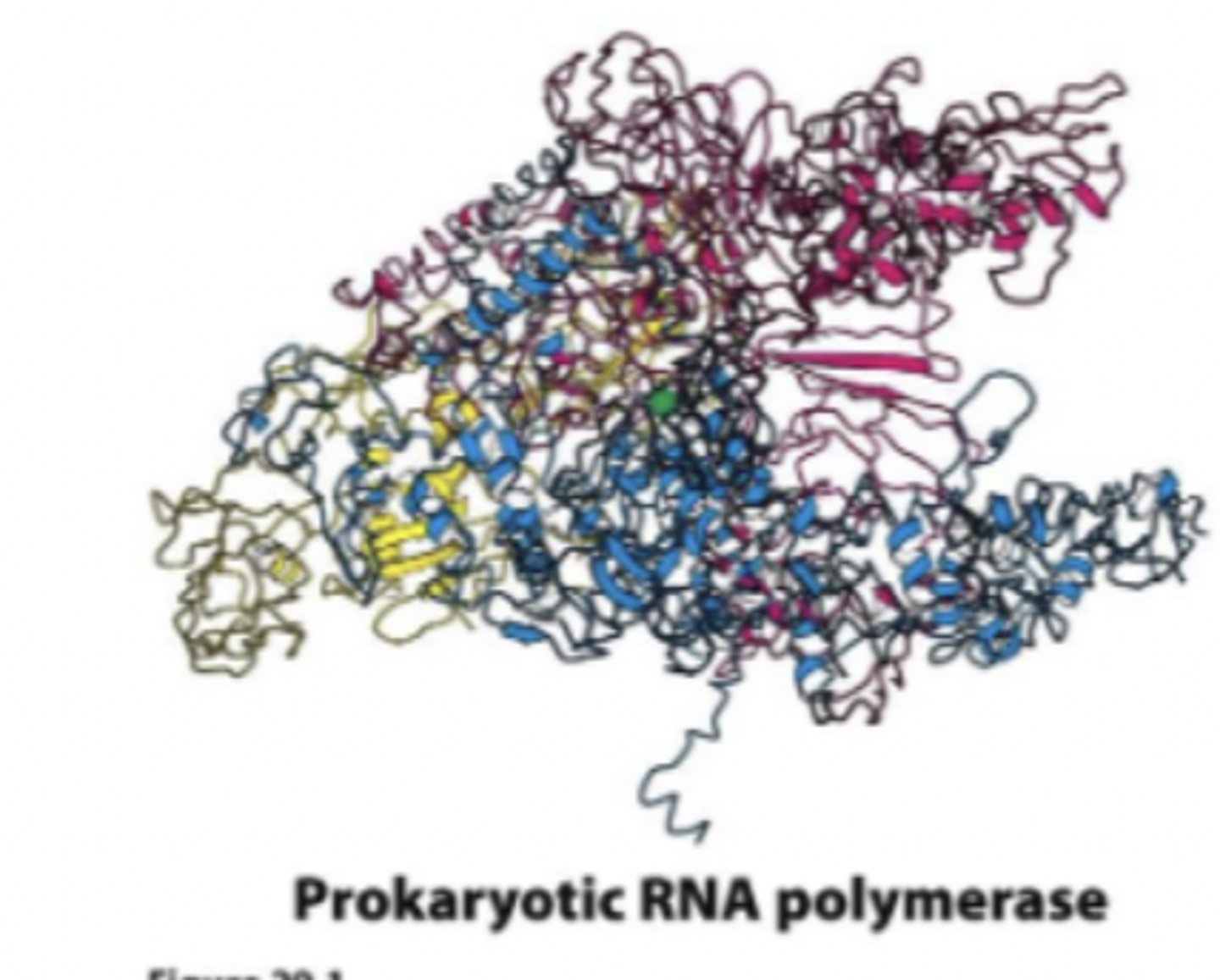
Prokaryotic RNA polymers
E. coli has a single polymerase
synthesizes all transcribed products (rRNA, mRNA, tRNA)
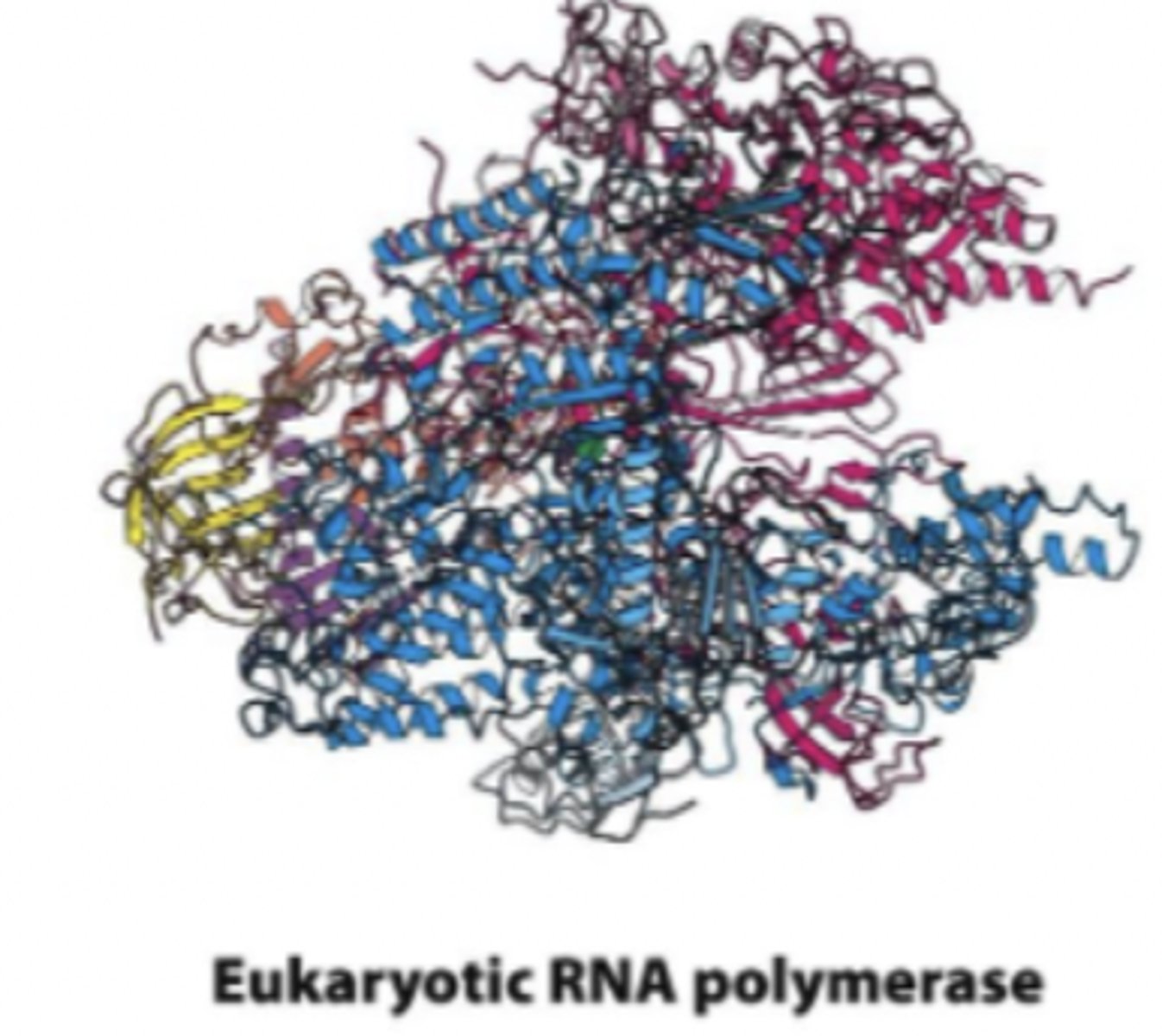
Eukaryotic RNA polymerase
humans have three (I, II and III)
different polymmyerases synthesize different RNA products (e.g., Pol II generates mRNA)
Important aspects of RNA synthesis
1. requires all four activated ribonucleotides and Mg2+ or Mn2+
2. New RNA strand has complementary sequence to template DNA strand
3. RNA polymerase does not require a primer
4. Errors generally not corrected by RNA polymerase; little proofreading

Prokaryotic RNA synthesis: Initiation - promoters
special DNA sequence located upstream of the start site
Prokaryotic RNA synthesis: Initiation
promoters direct RNA polymerase to transcription site
may contain 1 or more upstream promoter (UP) element
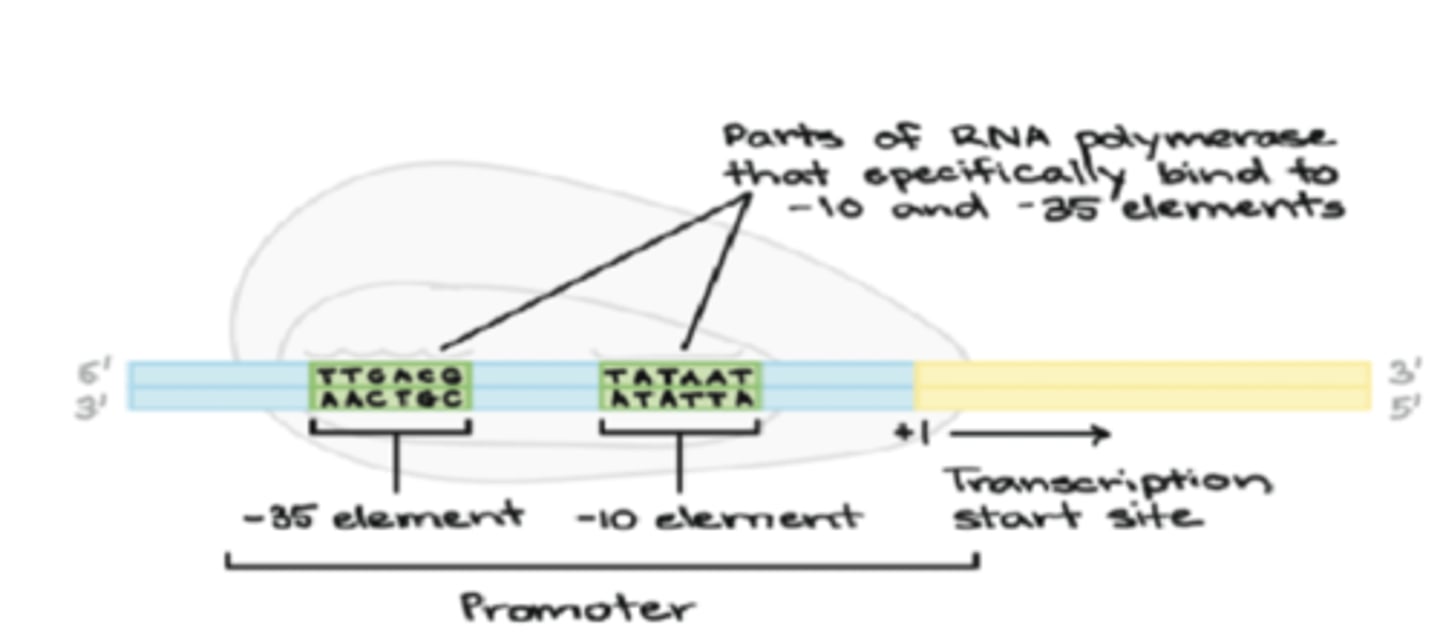
Prokaryotic RNA synthesis: Initiation site
in prokaryotes, known as -10 and -35
Prokaryotic RNA synthesis: Initiation - transcription factors
A DNA-binding protein; may also bind ligands (signal-sensing)
play an important regulatory role
ligands
molecule that binds another (generally larger) molecule
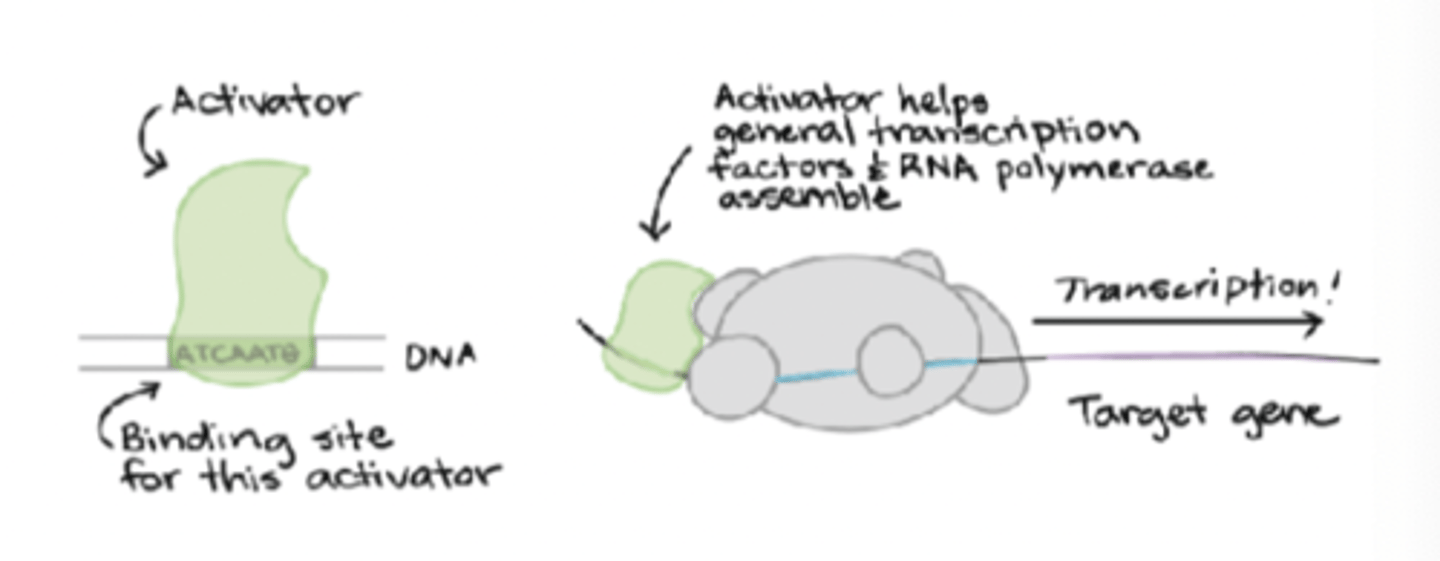
Prokaryotic RNA synthesis: Initiation - activator
if bound to a promoters upstream region, then activate transcription by recruiting RNA polymerase
Prokaryotic RNA synthesis: Initiation - repressor
if bound to a promoter, then suppress transcription by interfering with RNA polymerases access to start site
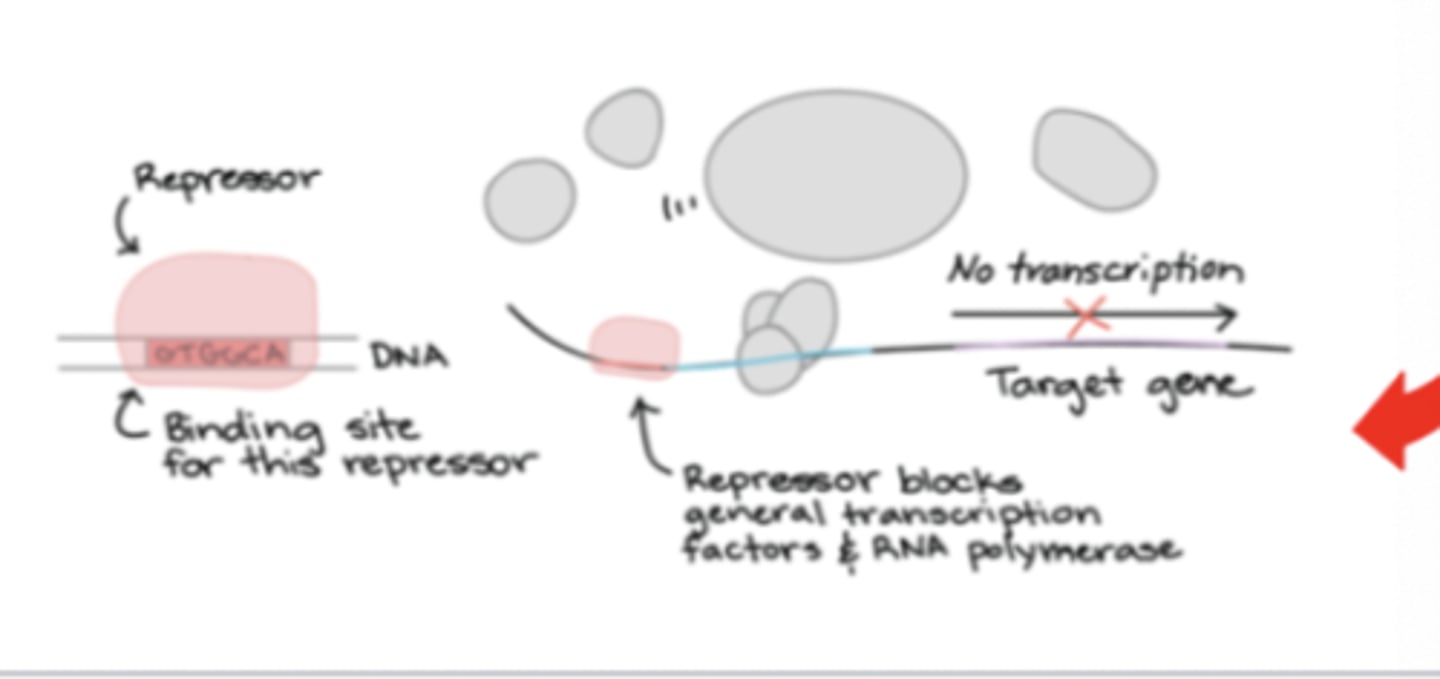
Prokaryotic RNA synthesis: Initiation - E. coli RNA polymerase
has 5 subunits (α2ββ'ωσ; holoenzyme)
Prokaryotic RNA synthesis: Initiation - sigma subunit
locating the promoter depends on this
- decreases enzymes affinity for general DNA regions
- allows RNA polymerase to "scan" for promoter along DNA
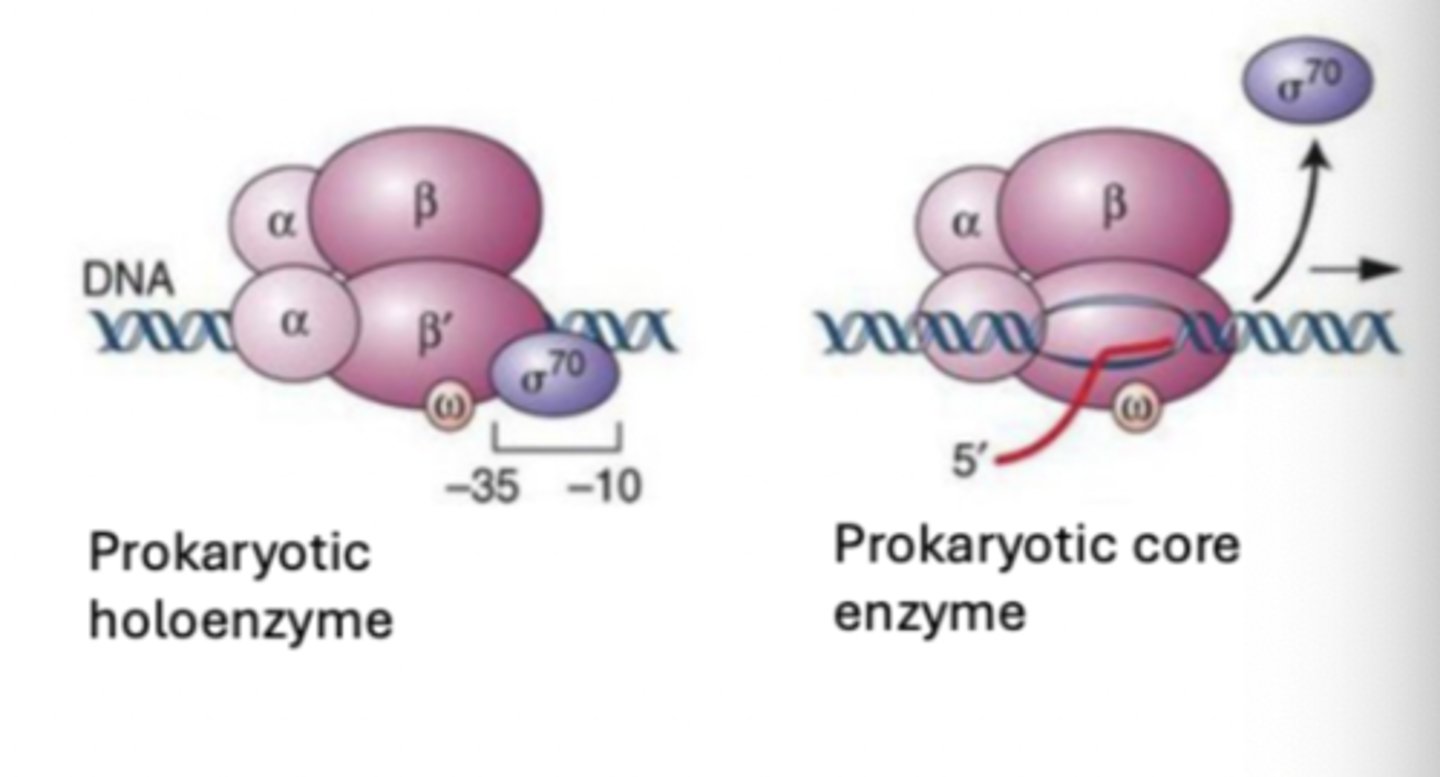
Prokaryotic RNA synthesis: Initiation - core enzyme
once located, sigma dissociates and ____ ______ (α2ββ'ω) remains
Prokaryotic RNA synthesis: Elongation
Core enzyme elongates RNA strand in a 5' to 3' direction
transcription bubble has polymerase, DNA template and nascent RNA
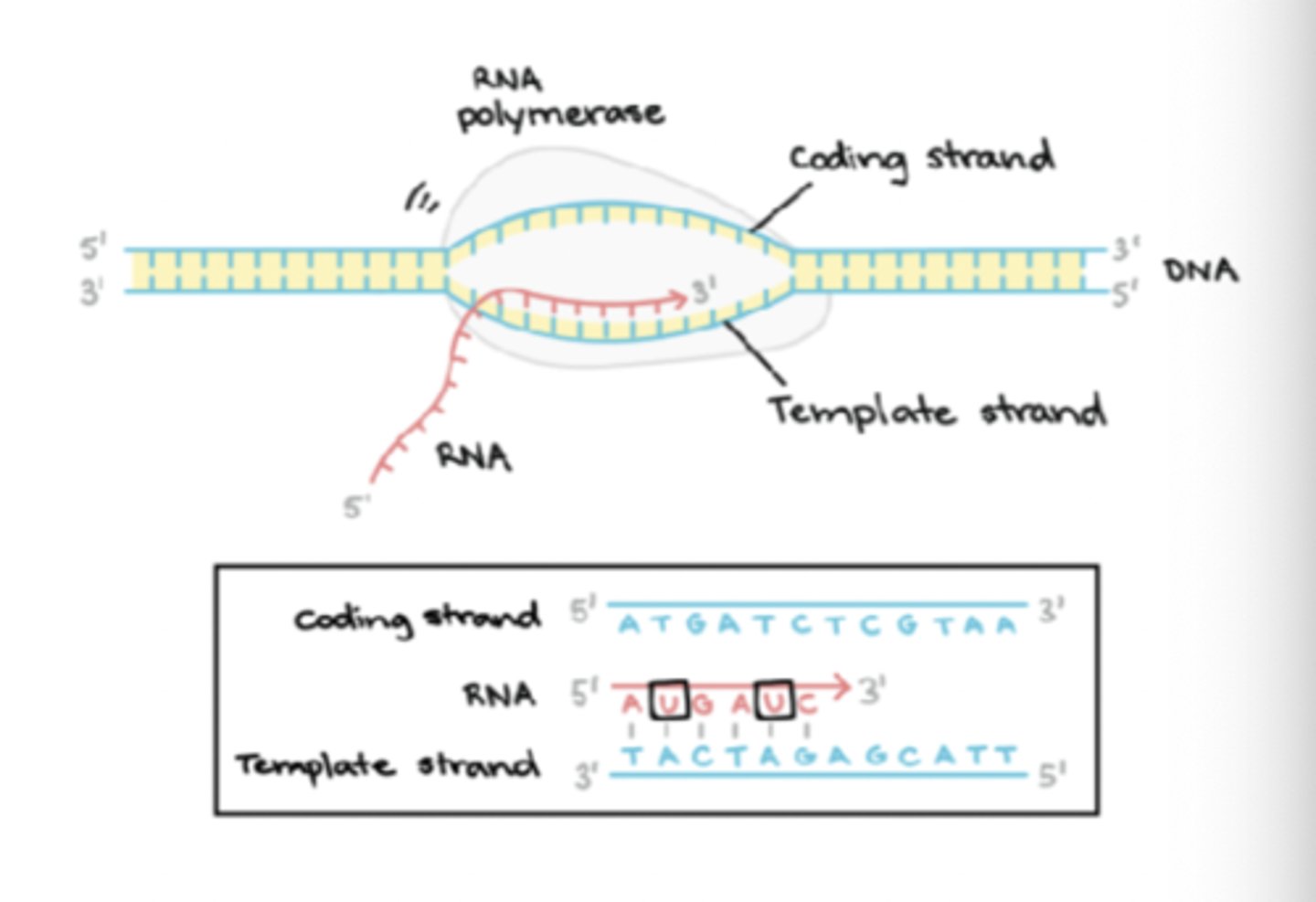
Prokaryotic RNA synthesis: Elongation - Nascent RNA
newly formed primary transcripts RNA
this forms a hybrid helix with ~8 base pairs of template DNA
Prokaryotic RNA synthesis - Termination
transcription terminated by synthesized RNA product of stop signals
not due to stop signals themselves within the DNA template
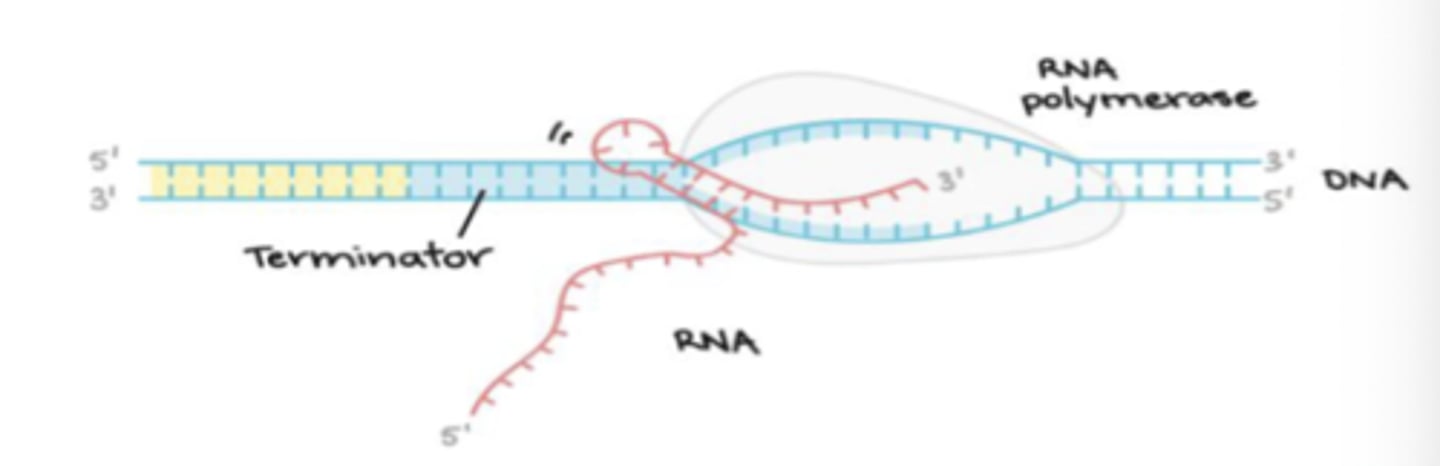
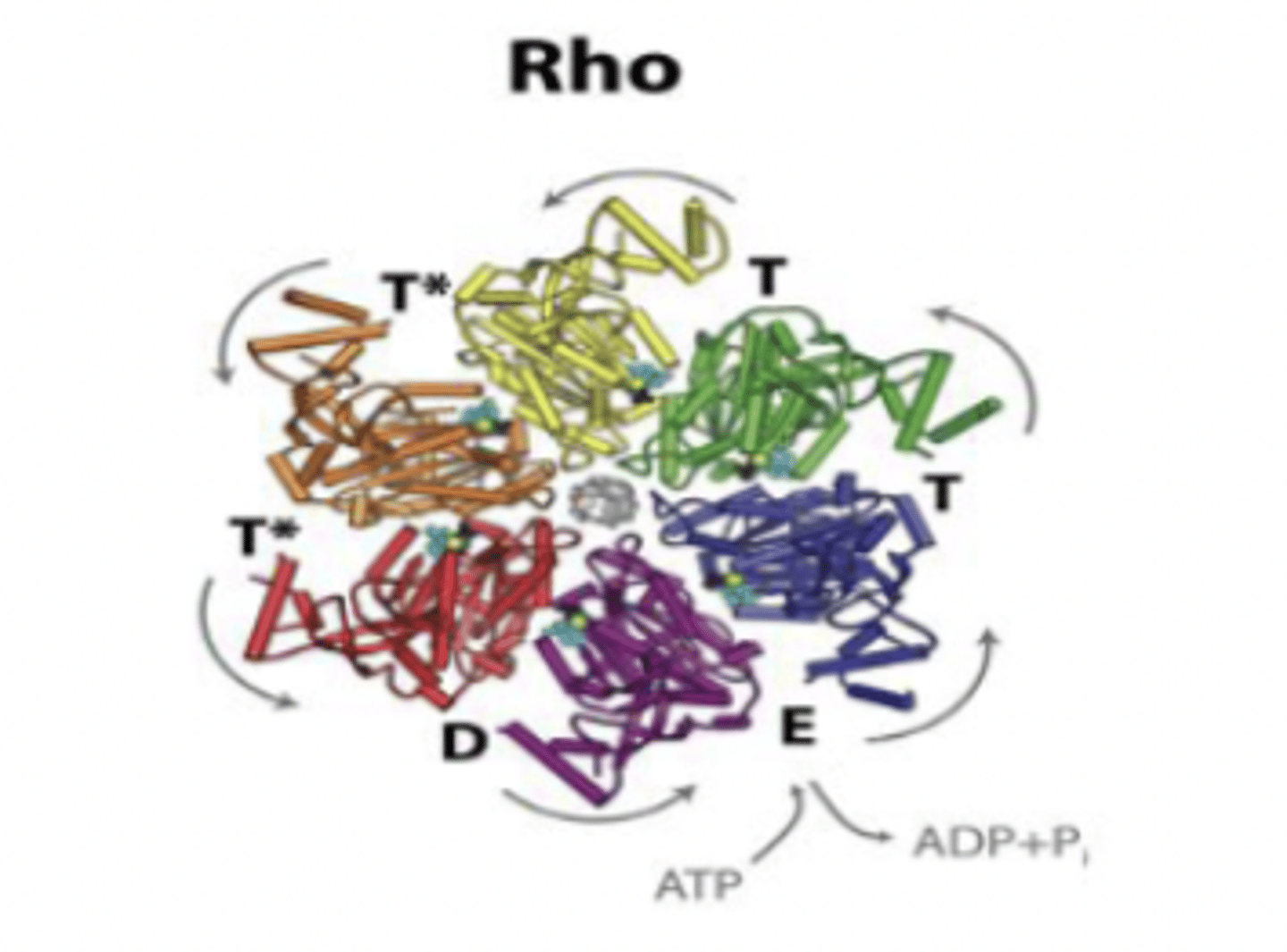
Prokaryotic RNA synthesis: Termination - Rho factor (p)
about 20% of prokaryotic termination events involve this factor
Rho factor (p)
an RNA binding protein that causes RNA polymerase to dissociate
Prokaryotic RNA Synthesis: RNA polymerase generated primary transcripts
Messenger RNA (mRNA)
Transfer RNA (tRNA)
Ribosomal RNA (rRNA)
Final products of prokaryotic RNA synthesis
RNA polymerases can generate three different primary transcripts
Nucleases
components of RNA synthesis (mRNA, tRNA, rRNA) excises by _________; may undergo further processing
How does eukaryotic RNA synthesis differ? - mechanistically
mechanistically similar in prokaryotic and eukaryotic organisms
How does eukaryotic RNA synthesis differ? genome size
genome size necessitates multiple promoters, with human genome estimated to have +9,000 across all chromosomes
Key differences in eukaryotic RNA synthesis
abundance of enhancer sequences
extent to which RNA product further processed / edited
abundance of "junk DNA"
Abundance of enhancer sequences - regulatory DNA sequences
these sequences (~50 - 1500 bp long) that are transcription factor binding sites
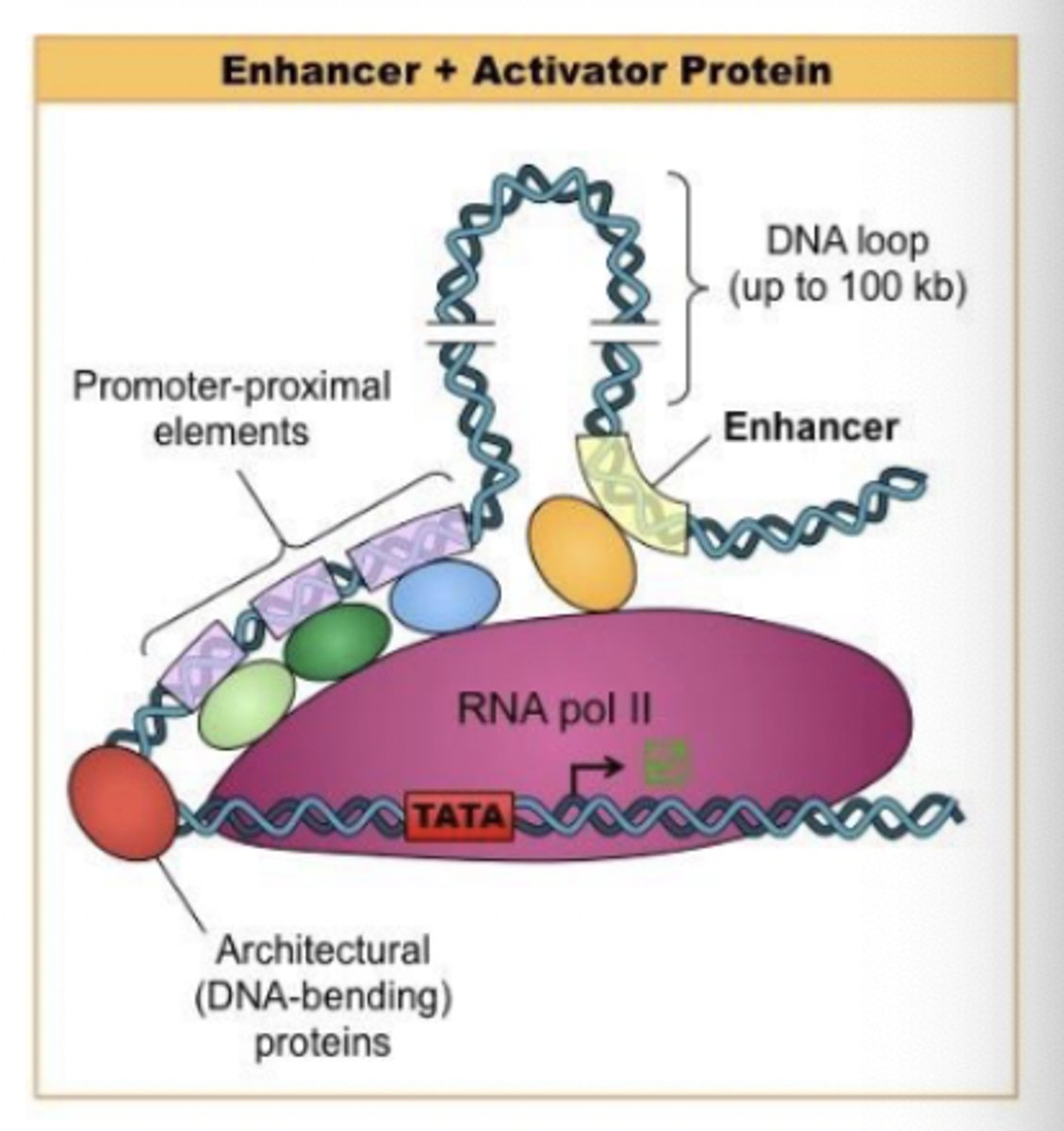
transcription factor binding sites
cause DNA to bend, bringing enhancer closer to promoter
help recruit RNA polymerase to the gene
Abundance of enhancer sequences
stimulate transcription over long distance (10^6 bp)
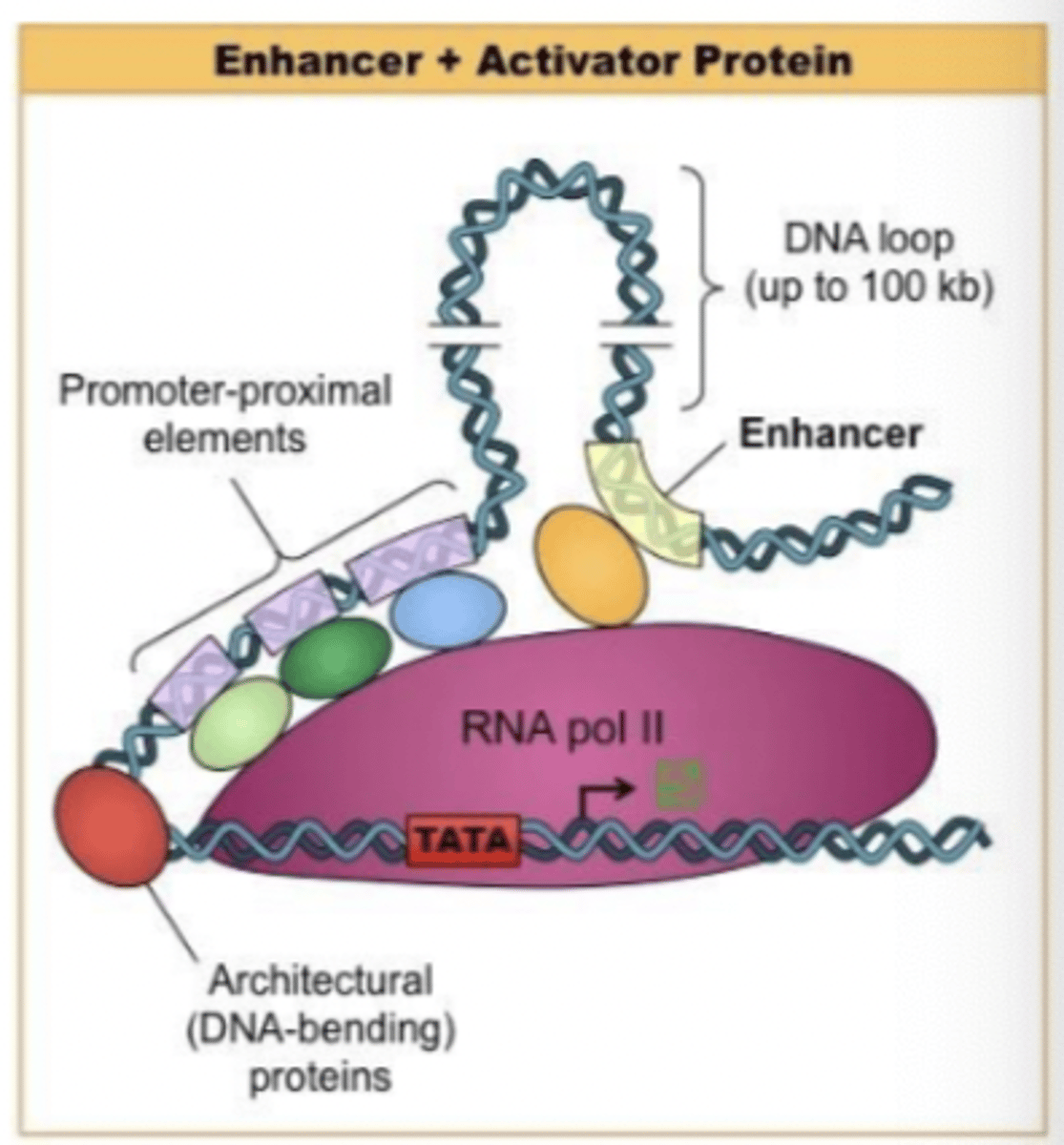
transcription over a long distance
may be upstream or downstream of gene
effective within introns or on either DNA strand
But must be within same chromosome as gene
enhancer sequences in prokaryotes
also in prokaryotes but much less abundant
intron
non-protein coding regions
RNA product further processed / edited
following intron removal, eukaryotic transcripts selectively methylated for stability and recognition
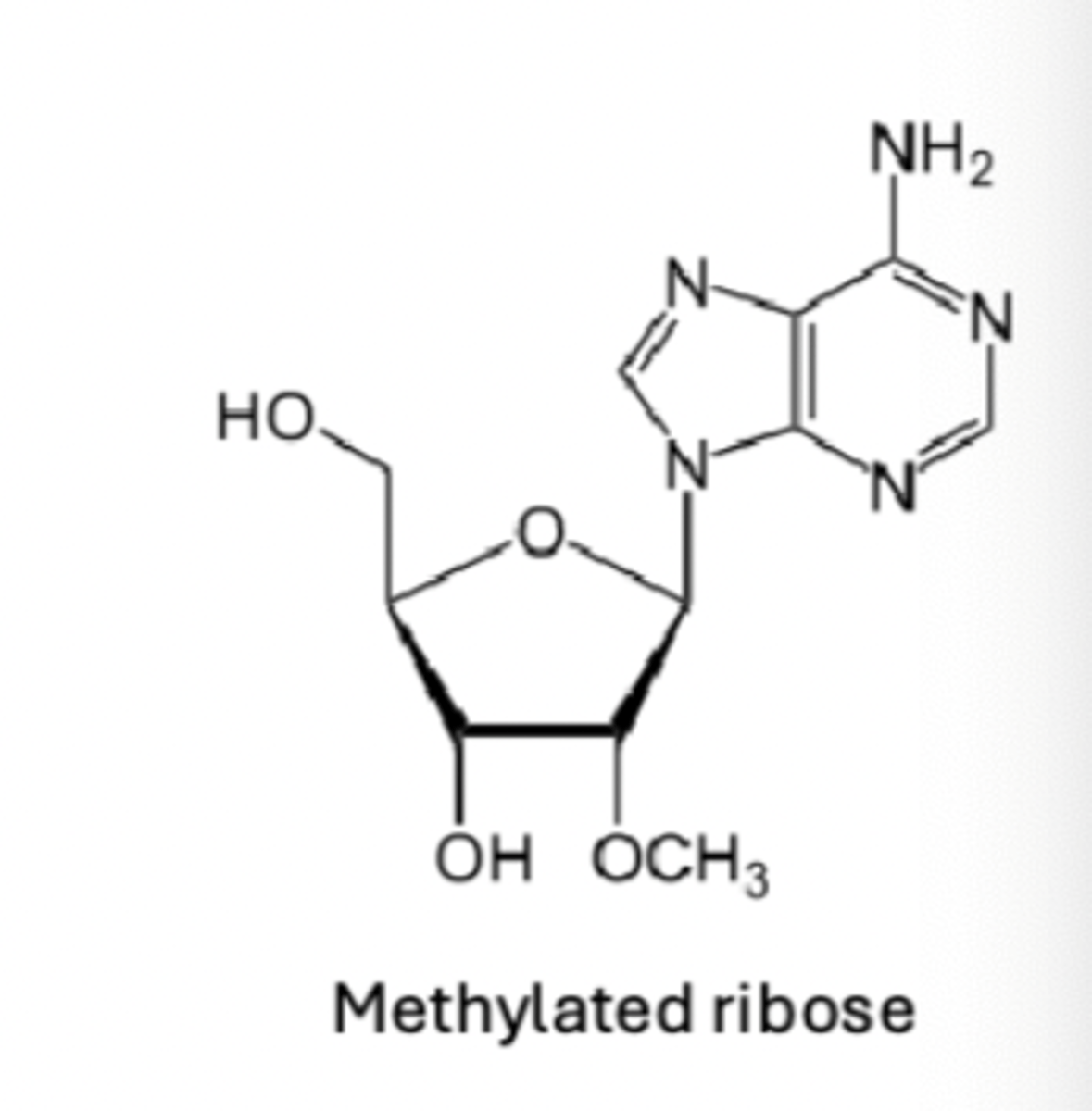
RNA product further processed / edited - mRNA transcripts
these also capped (5' end) and have a poly A tail added (3' end) to promote translation
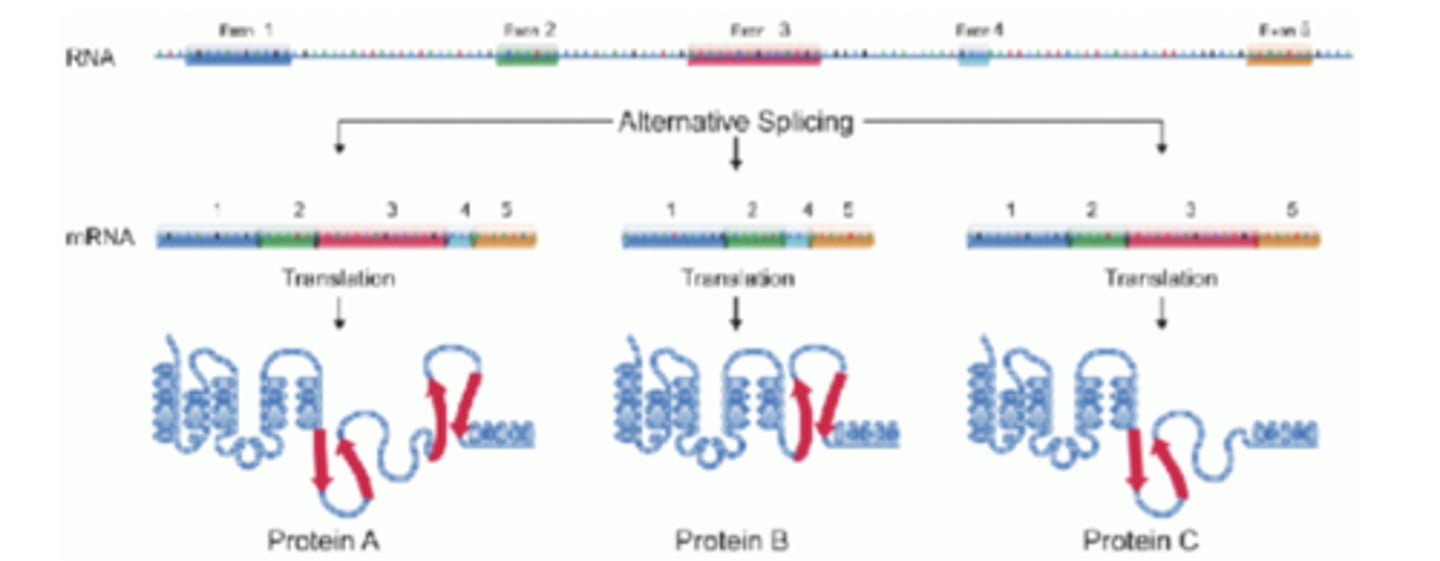
alternative splicing
this is used to increase proteome complexity
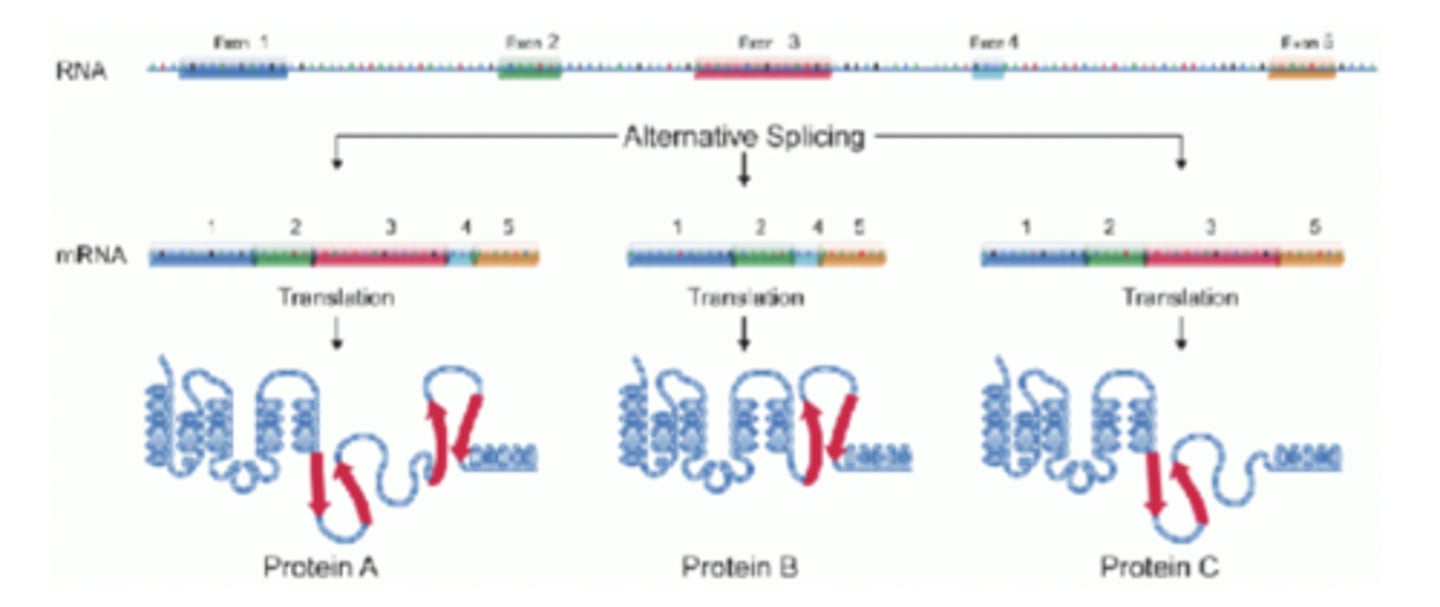
proteome
complete set of proteins expressed in a cell
Abundance of "junk DNA"
majority of eukaryotic DNA is non-protein coding
Why we have junk DNA
regulatory sequences (like promoters and enhancers)
protection from exonuclease (e.g., telomeres)
DNA that generates other transcribed products (tRNA and rRNA)
non-coding prokaryotic DNA
minority (<14%) of this DNA is non protein coding
mitochondrial genome
has 37 genes
mitochondrial 37 genes
13 for protein components of the electron transport chain and ATP synthase
22 for tRNA
2 for rRNA
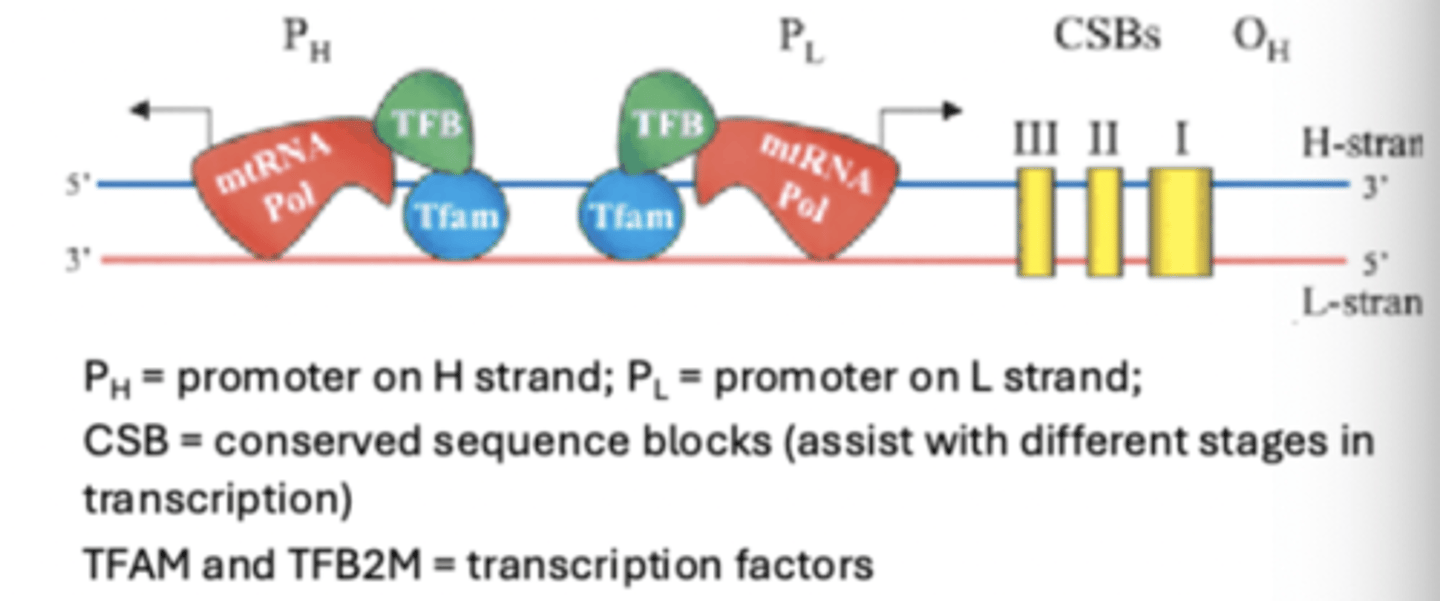
mitochondrial transcription
resembles prokaryotic process, as a single polymerase synthesizes all transcribed products but two
Two exceptions to what a single polymerase synthesizes in the mitochondria
- polymerase (POLRMT) is not multi-subunit (i.e., no sigma subunit)
- termination involved mTERF protein, which induces base flipping in DNA
true
true or false
RNA synthesis does not require DNA synthesis to happen beforehand
false
true or false
DNA is identical in all somatic cells, and gene expression is the same within the +200 different cell types
false
true or false
RNA polymerase has strong proofreading capabilities like DNA polymerase
E.) all of the above
RNA polymerase can generate a primary transcript containing
A.) rRNA
B.) tRNA
C.) mRNA
D.) none of the above
E.) all of the above