MLB 111 - Study Unit 7 Learning Outcomes
1/18
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
19 Terms
Define genotype, phenotype, allele, homozygous and heterozygous.
Genotype - the genetic makeup of an individual; the combination of alleles at a specific gene make up the genotype.
Phenotype - the expression of the genotype.
Allele - Different forms of a gene; correspond to different DNA sequences.
Homozygous - two copies of the same allele for a gene.
Heterozygous - two different alleles of the same gene.
Briefly explain how mutations can be either neutral, harmful, or beneficial.
Harmful - A mutation in the HTT gene increases glutamine amino acids in the protein product and results in Huntington’s disease.
Beneficial - Some adults can make lactase due to an enhancer mutation. This allows them to digest lactose.
Neutral (have no effect) - A neutral mutation in a human taste receptor gene removes the ability to taste a specific chemical, but it does not affect them.
Explain how environment can affect the effect of a mutation.
Some mutations may only be beneficial in certain regions.
Example: A mutation in a subunit of hemoglobin affects its shape. Individuals heterozygous for this have anaemia symptoms. In malaria area, these are more likely to survive malaria, providing a benefit in that environment.
Contrast the effect of somatic and germline mutations.
Somatic mutations occur in non-reproductive cells.
Germ-line mutations occur in reproductive cells and are passed on to offspring.
Germ-cell mutations - are transmitted to the next & future generations, somatic-cell mutations are not
The rate of mutation per nucleotide per replication is GREATER in SOMATIC cells than in germ cells because DNA repair mechanisms are more efficient in germ cells.
Cancer is a result of somatic mutation.
While somatic mutations can be devastating for the individual, germline mutations have a broader and potentially more severe impact due to their heritable nature, affecting future generations and potentially leading to widespread genetic disorders within a population.
Define a point mutation, and how it relates to a SNP
A point mutation is a change in a single nucleotide (nucleotide substitution).
A SNP (single-nucleotide polymorphism) is the result of a point mutation that occurred in the past and increased in frequency so that many individuals in the population now carry it.
Explain how the position of a mutation affects the effect it will have. Also refer to synonymous, nonsynonymous and silent mutations.
Point mutations in coding sequences have predictable consequences:
- Synonymous (silent): normal base pair substituted with another; normal codon becomes mutant codon, however both code for same amino acid and thus result in same amino acid sequences. If with a mutation there is no change in the amino acid in the protein, the mutation is a silent mutation.
- Nonsynonymous (missense): normal base pair substituted with another and the amino acid sequence changes, the change in mRNA codes for different amino acids. Can be harmful.
- Nonsense: creates a stop codon which prematurely terminates translation. Almost always harmful, truncated polypeptides are nonfunctional, unstable, & quickly destroyed.
Briefly discuss frameshift mutations and the DNA changes that results in such mutations.
An insertion/deletion of some number of nucleotides in the DNA sequence that is NOT a multiple of three causes a shift in the reading frame of the mRNA, changing all following codons.
An insertion or deletion of three bases maintains the reading frame but inserts or deletes one amino acid from the translated protein.
Briefly mention the role of transposable elements in mutation.
Transposable elements are DNA sequences that can replicate and move from one location to another in a DNA molecule.
When such a large piece of DNA inserts into a gene it interferes with transcription, causes errors in RNA processing, or disrupts the open reading frame.
Briefly discuss chromosomal mutations by referring to duplications and deletions of chromosome sections.
Chromosomal mutations delete/duplicate regions of a chromosome.
These mutations alter the linear order of genes along a chromosome, interchange arms of nonhomologous chromosomes, and affect chromosome pairing & segregation in meiosis.
Duplication: region of a chromosome that is present twice instead of once. Duplication of a region of the genome is typically less harmful than deletion of that region.
Deletion: a missing region of a gene or chromosome. Deletions can arise from a replication error or joining of double-stranded breaks during DNA repair.
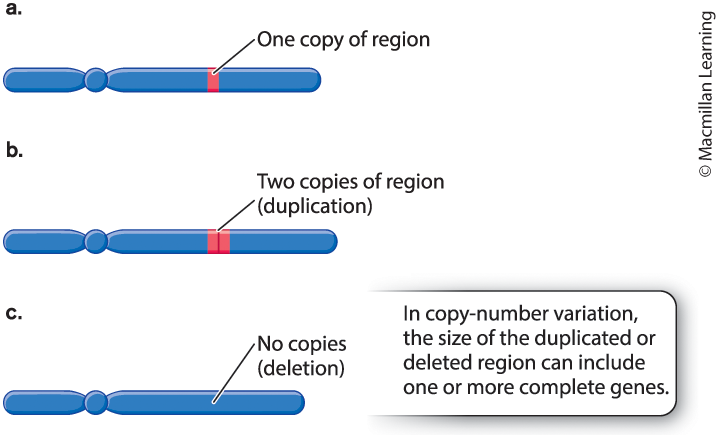
Define copy-number variations (CNV) and their role in creating genetic variation. Also, refer to tandem repeats and their use in DNA typing.
CNV, differences among individuals in the number of copies of a region of the genome.
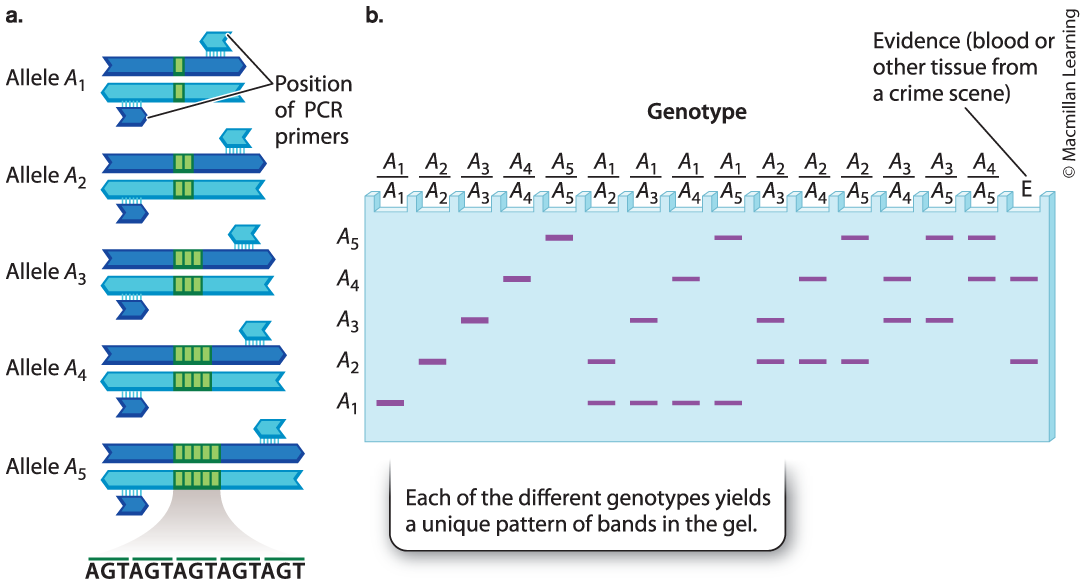
Tandem repeat: a region along a DNA molecule in which many identical copies of a short sequence of nucleotides are adjacent to one another.

Tandem Repeats
Tandem repeats are useful in DNA typing - the analysis of a small quantity of DNA to uniquely identify an individual; also called DNA fingerprinting.
The distinct patterns of bands can be used to identify the genotype of any individual for a particular region of DNA. (criminal identification)
Briefly discuss chromosomal inversions and translocations as sources of DNA variation.
Inversion is the reversal of the normal order of a block of genes; typically produced when the region between two breaks in a chromosome is flipped in orientation before the breaks are repaired. Large inversions cause problems with meiossis. Small inversions are common and have an important role in chromosome evolution. Accumulation of inversions explains why the order of genes along a chromosome can differ among closely related species.
Reciprocal translocation is the interchange of parts between nonhomologous chromosomes. Both chromosomes are broken and terminal segments are exchanged before the breaks are repaired. Do not affect survival. If both chromosomes involved in reciprocal translocation DON'T move together into same daughter cells, resulting gametes have one part of the reciprocal translocation. Gametes have extra copies of genes on one chromosome, and are missing copies of genes in the other.
Define mutagens and their role in generating mutations.
Mutagens: an agent that increases the probability of a mutation. Many different types of mutations are spontaneous and caused by mutagens.
Mutations can be caused by X-rays; UV light; hydrogen peroxide; highly reactive chemicals; and radiation.
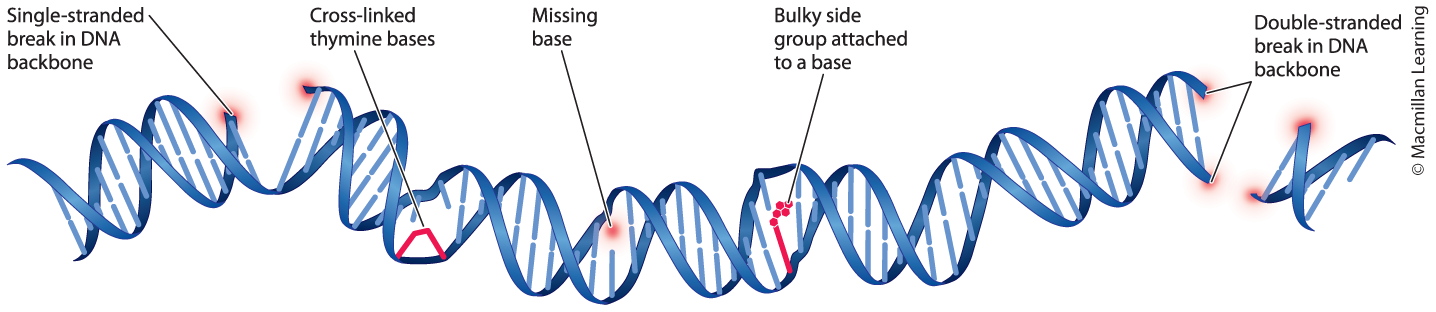
Damage induced by mutagens can affect the structure of the DNA double helix.
- Single-stranded break in DNA backbone
- Cross-linked thymine bases
- Missing base
- Bulky side group attached to a base
- Double-stranded break in DNA backbone
DNA Repair
There are many different repair mechanisms to restore DNA to its original sequence.
Post replication mismatch repair
Base excision repair
Nucleotide excision repair
Post replication Mismatch Repair
Corrects replication errors and other base mismatches.
Mismatch Recognition: Mismatched bases can be recognized by the change in the shape of the DNA double helix. The first stage of mismatch repair involves breaking the sugar-phosphate backbone.
Replacement of Mismatched Bases
Base Excision Repair
Removes base adducts, uracil, abasic sites and oxidative lesions.
Removal of Chemically Modified Bases: Base excision repair is different from nucleotide excision repair because, in nucleotide excision repair, the sugar-phosphate backbone is broken first and then an exonuclease removes many bases.
Addition of the Appropriate Nucleotide: Once the base is removed, the sugar-phosphate backbone is opened and the sugar is removed. The other strand serves as a template, so the correct base can be added. DNA ligase will make the sugar-phosphate backbone continuous to complete the repair.
Nucleotide Excision Repair
Removes bulky lesions and intrastrand crosslinks.
Removing Multiple Bases: Nucleotide excision repair can remove multiple mismatched bases or bulky lesions, like thymine dimers caused by UV damage.
DNA ligase will make the sugar-phosphate backbone continuous to complete the repair.
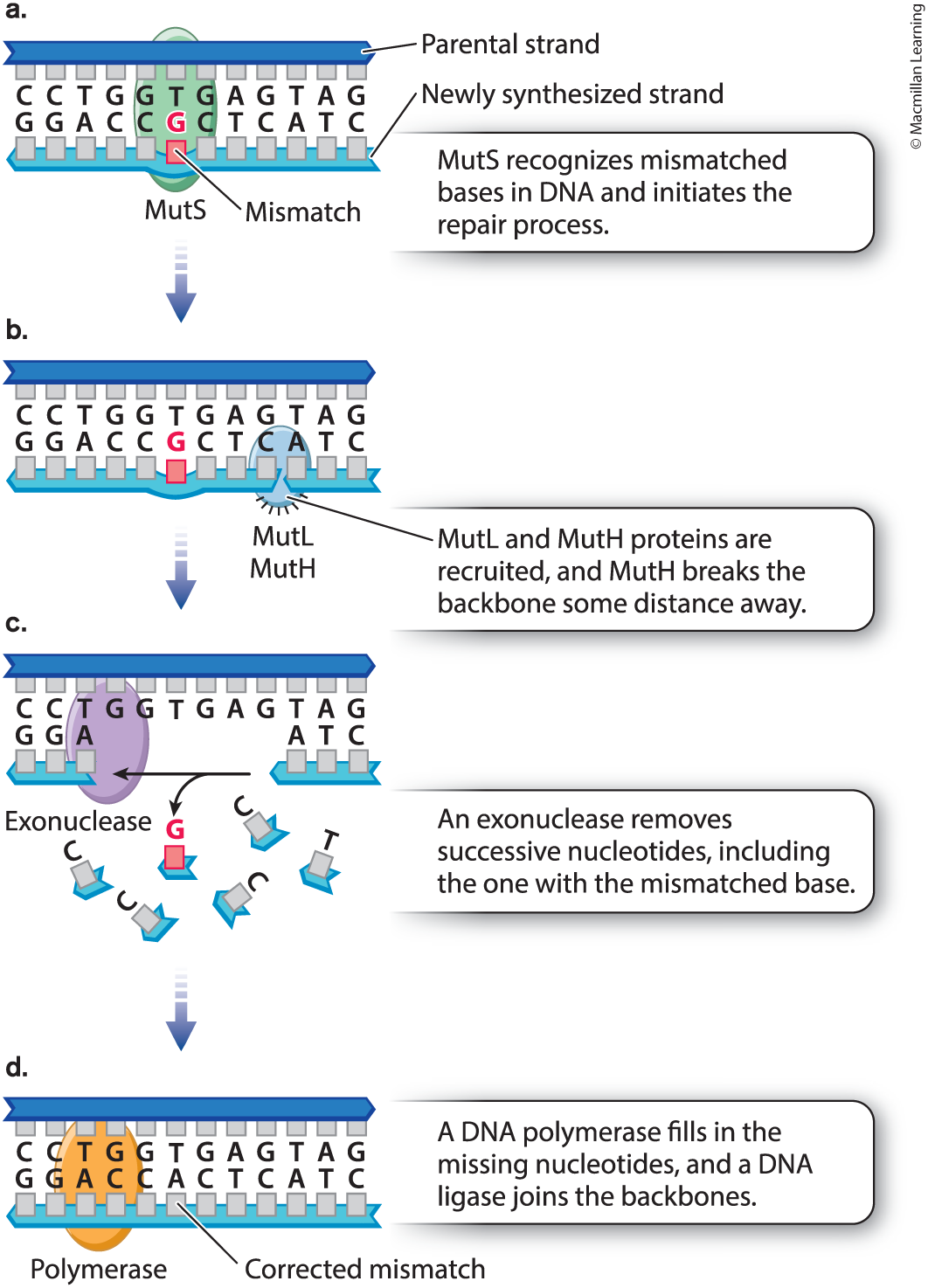
Discuss mismatch repair in E. coli. You do not need to know the individual components, but rather the overall process.
Protein (MutS) recognizes and binds to site of mismatch & brings two other proteins (Mutt & MutH) to site. Next protein (MutL) determines which DNA backbone will be cleaved. Third protein (MutH) cleaves backbone in vicinity of mismatch. Exonuclease degrades cleaved strand to a distance beyond site of mismatch. DNA polymerase synthesises new DNA to fill gap & thus corrects mismatch. DNA ligase seals remaining nick in DNA backbone. Daughter strand usually cleaved.
Contrast the mechanisms and functions of base excision repair and nucleotide excision repair.
Nucleotide excision repair is similar to mismatch repair, with the difference being the number of mismatched bases. Nucleotide excision repair can recognize groups of adjacent nucleotides that are mismatched or damaged, whereas mismatch repair can recognize only single mismatched bases.