Virus
1/57
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
58 Terms
Viruses are
simplest biological systems, they are infectious particles consisting of 1) nucleic acid, 2) capsid, and 3) in some cases, a viral envelope
Describe the structure of capsid protein
it is a protein coat, built from a large number of protein subunits called capsomeres.
there are many capsid shapes (eg. helical, icosahedral, complex)
viral nucleic acid + its surrounding protein capsid is known as a nucleocapsid
What are the functions of capsid proteins
encloses the viral genome and protects it from digestion by enzymes.
aids in attachment to and penetration of the host cell
carries viral enzymes involved in viral replication
Describe the structure of viral envelopes
contains of a phospholipid bilayer (derived from the host’s plasma membrane during budding) which surrounds the capsid
some contain glycoproteins that project from the viral envelope as spikes
glycoproteins are encoded by the viral genome (eg hiv)
State the function of viral envelope
glycoproteins on the viral envelope are complementary in shape to and can therefore bind to the receptor proteins on the host cell membrane, determining the specific host range (i.e. specific cell types or species/organisms that a virus can infect)
some viruses have broad host ranges (E.g. West Nile virus) while some viruses have narrow host ranges (E.g. HIV)
viral envelope facilitates the entry of viruses into host cells by either direct fusion with the host cell membrane or through receptor-mediated endocytosis
Virus exhibit
both living and non-living characteristics.
How do viruses attach to host cell membrane?
proteins/glycoproteins on viral capsid/outer envelope have shapes complementary to specific receptors onbthe host cell membrane and bind to them
How do animals virus enter host cell?
viruses then enter the host cell either via direct fusion between the viral envelope and host cell surface membrane or receptor-mediated endocytosis
viral DNA/RNA and capsid proteins are released during un-coating
How does virus synthesise?
host or viral enzymes to synthesise new copies of viral genome (replication)
host enzymes and ribosomes to express viral genes, producing viral proteins
Describe positive viral mRNA
considered as viral mRNA
directly translated into viral proteins
Describe negative viral mRNA
nucleotide sequence that is complementary to that of viral mRNA / (+) strand viral RNA
cannot be directly translated into proteins
must be converted to (+) RNA by a viral RNA polymerase prior to translation
Describe the structure of T4 bacteriophage
complex virus with an icosahedral capsid attached to a tail apparatus.
tail apparatus is a contractile tail, composed of an inner core (allows injection of viral DNA) and an outer contractile sheath (contracts during infection of bacteria)
at the end of the tail is a baseplate, with attached tail fibres and tail pins. The base plate is the point of attachment to its host cells
T4 phage genome is a linear, double-stranded DNA, thus, it is a DNA virus.
Describe attachment for T4 bacteriophage
tail fibres of the phage are complementary in shape to the specific receptor sites on the host’s cell surface and bind to them.
upon attachment, lysozymes are released from the tip of base plate to degrade a portion of the bacterial cell wall, inserting of its tail core
Describe penetration for T4 bacteriophage
cntractile sheath of the tail contracts and drives the hollow core through the
bacterial cell wall and membrane.
causingthe injection of phage DNA into the host
empty capsid remains outside the host cell
Describe synthesis for T4 bacteriophage
synthesis of early proteins occur first
one of the first phage genes expressed codes for a phage enzyme (DNase) that degrades the host cell’s DNA
shuts down the synthesis of the bacterium’s DNA, RNA, and proteins
pther phage genes expressed code for proteins that are needed to replicate the phage DNA
synthesis of late proteins occur next
newly replicated copies of phage DNA is used for transcription and translation to make structural proteins
phage lysozyme is also made and packaged into the tail of the phage (to allow it to escape from the host cell during the last step)
Describe assembly and realise for T4 bacteriophage
phage proteins are synthesised, they self-assemble to form the capsid components (head, tail, tail fibres)
phage DNA is packaged into the capsid (encapsidation) as the head forms.
phage lysozymes are produced, and digest the host’s cell wall
damaged cell wall enables entry of water into the host bacterium cell via osmosis, causing it to swell, lyse and release numerous completed T4 bacteriophages to infect other cells
Describe the structure of lambda phages
lambda () phage is a complex virus with an icosahedral capsid and a non-contractile tail sheath consisting of a single tail fibre
lambda phage genome is a linear, double-stranded DNA which can circularize in the host cell
it is a DNA virus
lambda () phage is a temperate phage as it is able to reproduce by lysogenic and lytic cycles
Describe adsorption of lambda phage
tail fibre of lambda () phage has a shape complementary to the specific receptor site on the host’s cell surface and binds to it
Describe penetration of lambda phage
the phage then injects its DNA into the cytosol of the host cell, within the host, the phage DNA circularizes
Describe genetic integration of lambda phage
circularized DNA integrates into a specific site on the host bacterial chromosome
done by viral proteins that break both circular DNA molecules and join them to each other
integrated viral DNA is known as a prophage.
resulting host cell containing the prophage is known as the lysogen.
prophage stage, one prophage
gene codes for a repressor protein that prevents the transcription of most of the remaining prophage genes.
phage genome is mostly silent within the bacterium
Describe replication of lambda phage
every time the host cell prepares to divide by binary fission, the phage DNA is replicated along with the bacterial chromosome and is passed on to daughter cell
single infected cell can quickly rise to a large population of bacteria carrying the virus in the form of prophages
this mechanism enables viruses to propagate without killing the host cells
Describe induction of lambda phage
stressful environmental signal (e.g. High-energy radiation, presence of certain toxic chemicals, lack of nutrients etc.) can trigger induction whereby the virus switches its replication mode from lysogenic to lytic
when this occurs, the prophage is excised from the host bacterial chromosome certain proteases are produced, hydrolysing the repressor protein, allowing the remaining prophage genes to be expressed
results in the start of the lytic cycle
State the difference between lytic and lysogenic cycle (effect on host genome)
lytic: host cell’s DNA is hydrolysed by phage DNA, phage DNA remains in cytoplasm
lysogenic: host cell’s DNA is not hydrolysed by phage DNA, phage DNA remains in cytoplasm
State the difference between lytic and lysogenic cycle (effect on host genome)
lytic: phage DNA exploits host machinery to replicate phage DNA and proteins
lysogenic: phage DNA (prophage) replicates when host DNA replicates before host cell divides
State the difference between lytic and lysogenic cycle (replication of phage DNA)
lytic: phage DNA exploits host machinery to replicate phage DNA and proteins
lysogenic: phage DNA (prophage) replicates when host DNA replicates before host cell divides
State the difference between lytic and lysogenic cycle (release of new phages)
lytic: phage DNA and proteins assemble to form completed phages, phage lysozyme weakens host cells wall, resulting osmosis cause lysis of host to release new phages
lysogenic: prophage not released. Remains integrated with host DNA and does not exit host until it enters lytic mode
State the difference between lytic and lysogenic cycle (death of host cells)
lytic: occurs when cell lyses to release new phages
lysogenic: phage genome replicates without destroying host cell
Describe the structure of influenza virus
viral envelope (derived from the phospholipid bilayer of the plasma membrane of the host cell during budding) surrounds eight helical nucleocapsids, each made up of nucleocapsid proteins associated with viral RNA, virus appears spherical in shape
viral envelope has embedded glycoproteins
haemagglutinin (HA): For binding of viruses to sialic acid / neuraminic acid-containing
receptor sites on the surface membrane of target host cells (i.e. involved in viral entry)
neuraminidase (NA) (aka sialidase): An enzyme that cleaves sialic acid / neuraminic acid
residues from the host cell surface, allowing the budding viruses to be released from infected cells (i.e. involved in viral release)
HA and NA can serve as targets for antiviral drugs
viral genome consists of eight segments of linear (-) single-stranded RNA wrapped around nucleocapsid proteins (also called nucleoproteins)
end of each RNA segment is attached to an RNA-dependent RNA polymerase
these viral RNA-dependent RNA polymerases use the (-) strand RNA as a template to synthesise the complementary (+) strand RNA
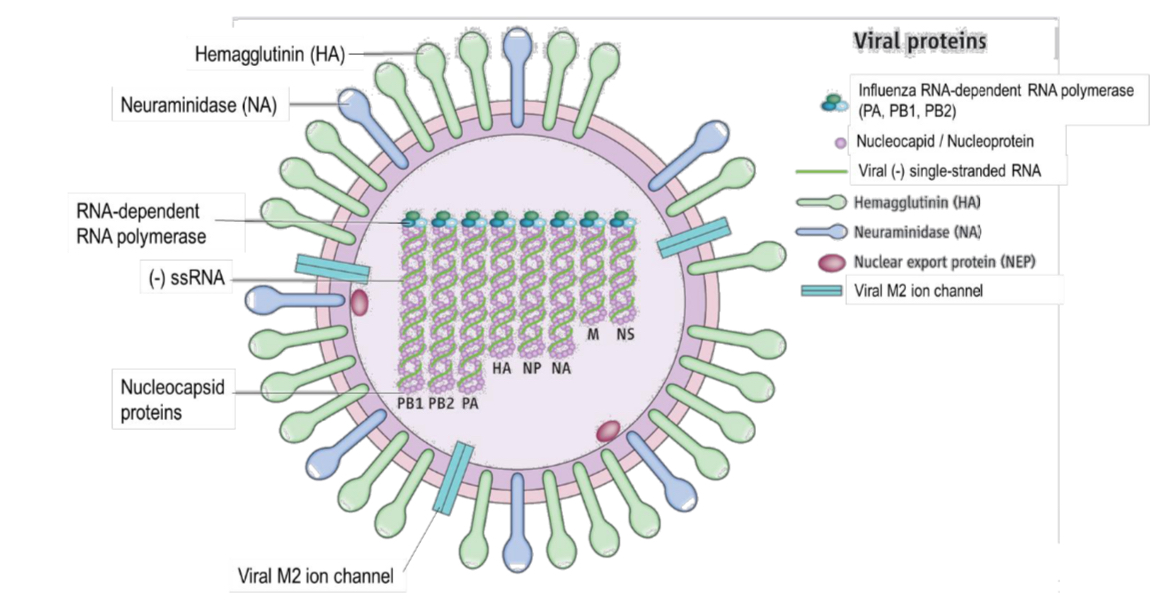
What is a influenza virus
Influenza is an infectious disease affecting mainly birds and mammals
symptoms of disease: Fever, cough and severe muscle aches
can cause severe illness or death of people at high risk
Influenza viral proteins trigger the body’s immune response, causing inflammation of the epithelial linings of the airways,
Inflammation results in symptoms of the disease
in severe cases, lung inflammation may develop into pneumonia (lung infection), as the airways are blocked by fluid and there is reduced clearance of infectious agents, result in death.
What is the mode of transmission for influenza virus
commonly spread through aerosols
What are the target cells of influenza viruses
target cells: Influenza viruses infect the epithelial cells of the respiratory system (e.g. nose, throat, lungs of mammals)
What does type A primary inflect
A strains cause severe illness and can infect people, birds, pigs and other animals, only type to have caused human pandemics (global disease outbreak).
within this strain, the influenza viruses are further categorized into subtypes based on type of glycoproteins on the viral envelope, namely haemagglutinin (HA)
What does type B primary inflect
B strains are normally found in humans and cause sporadic small scale outbreaks. These viruses are not classified into subtypes.
What does type C primary inflect
C strains cause mild illness in humans and no outbreaks in the population, not classified into subtypes.
What does type D primary inflect
D strains primarily affect cattle. They are not known to infect or cause illness in people.
Describe the attachment of influenza
haemagglutinin glycoproteins (HA) on the viral envelope recognize and bind to specific, complementary receptor sites (containing sialic acid / neuraminic acid residues) on the surface membrane of the host epithelial cells of the respiratory system
Describe the penetration of influenza
virus then enters the host cell via receptor-mediated endocytosis, where the host cell membrane forms an endosome / endocytotic vesicle around the virus
endosome becomes acidic, and this environment triggers a conformational change in haemagglutinin protein, leading to the fusion of the viral envelope with the endosomal membrane
M2 ion channel found in the viral envelope is important for also acidifying the interior of the virus
triggers the release of individual viral nucleocapsids (from the matrix protein) into the cytosol.
cellular enzymes will then digest the viral nucleocapsid proteins to release the RNA segments (process is known as uncoating)
viral RNA genome / RNA segments then migrate to the nucleus
Describe the synthesis of influenza virus
the host nucleus, the viral (-) single-stranded RNA functions as a template for making complementary viral (+) strand RNA. This process is catalysed by the viral RNA- dependent RNA polymerase which is absent in the host cell
viral (+) strand RNA (viral mRNA) then acts as a template for:
replication of new viral (-) single-stranded RNA genome within the nucleus. This process is also catalysed by viral RNA-dependent RNA polymerase
translation into viral proteins (e.g. nucleocapsid proteins / capsomeres and envelope glycoproteins) using host machinery (e.g. ribosomes). This happens after the (+) RNA strands translocate out of the nucleus into the host cytoplasm
Describe the assembly and release of influenza virus
nucleocapsid proteins and RNA-dependent RNA polymerases produced in the cytoplasm are transported back into the host nucleus.
host nucleus, the nucleocapsid proteins and RNA-dependent RNA polymerases will associate with each viral (-) RNA segment to form RNA-nucleocapsid protein complexes, this is known as encapsidation.
newly-assembled RNA-nucleocapsid protein complexes migrate out into the cytoplasm and then towards the host cell membrane, where the glycoproteins HA and NA are incorporated
virus buds off (Reject: “exocytosis”) from the host cell, it is enclosed by host’s plasma membrane (consisting of the viral glycoproteins), which then forms the viral envelope
neuraminidase is then able to play the final role in virus budding – cleaving the sialic acid / neuraminic acid residues from the host cell surface membrane receptors, thus releasing the new viral particles from the infected host cells
budding itself does not necessarily kill the host cells (as opposed to lysis in the lytic cycle of phages), but eventually, the host cell will die due to depletion of energy and resources
enveloped viruses are now free to infect other epithelial cells
Describe HIV
HIV infection represents the early stages of infection of an individual by HIV and is a progressive disease. As HIV gradually attacks the immune system, a weakened immune system results, leading to acquired immunodeficiency syndrome (AIDS)
What are the target cells of HIV
CD4 T lymphocytes (i.e. white blood cells) and Macrophages
Why is HIV hard to eliminate
transcriptase, which serves to form double-stranded viral DNA from (+) single-stranded RNA viral genome. It is able to do so as the reverse transcriptase consists of the following activities
RNA-dependent DNA polymerase activity → forms single-stranded complementary DNA (cDNA) from a (+) single-stranded RNA template.
RNAse activity → removes single-stranded RNA from the viral RNA-DNA hybrid
DNA-dependent DNA polymerase → uses the single-stranded cDNA as template to
form double-stranded viral DNA.
Reverse transcription is prone to errors and the enzyme reverse transcriptase does not proofread.
errors and new mutations are introduced during this process.
reason why HIV is difficult for the immune system to eliminate
What is a retrovirus
“Retro” means backwards which refers to the reverse direction in which genetic information flows for these viruses (i.e. from RNA to DNA)
HIV carries an enzyme called reverse
(e.g. shape of viral surface proteins (e.g. gp120) is altered, so the antibodies that are made
against the earlier strains of HIV can no longer bind)
What are the symptoms of HIV
opportunistic infections (E.g. pneumonia) caused by other pathogens due to a weakened immune system
replication of HIV in CD4 / helper T lymphocytes eventually kills the T lymphocytes (e.g. by triggering apoptosis)
as more HIV particles are released into the blood and more T lymphocytes are infected, T lymphocytes decrease in number, thus compromising the immune system
B lymphocytes produce antibodies with the help of T lymphocytes. With the decrease in T lymphocytes, less antibodies are produced to bind to pathogens (e.g. bacteria) to facilitate their destruction
What are the modes of transmission for HIV
direct exposure of a person’s blood to body fluids containing the virus (blood, semen, vaginal secretions) (e.g. sexual contact or contaminated needles).
infected mother to her baby via breastfeeding or during childbirth
Describe the structure of influenza
HIV consists of an envelope (derived from the phospholipid bilayer of the plasma membrane of the host cell during budding)
envelope has embedded glycoproteins (spikes) gp120 and gp41, both are involved in viral entry
envelope surrounds a cone-shaped capsid, which in turns encloses:
2 copies of linear, (+) single-stranded RNA associated with nucleocapsid proteins.
2 copies of reverse transcriptase.
other viral enzymes, integrase and protease
Describe attachment of HIV
once HIV enters the bloodstream, it circulates throughout the body. However, it only infects susceptible host immune cells
gp120 of HIV binds specifically to a complementary host cell surface receptor protein, CD4 (involved in immune recognition). CD4 is present on the surface
membrane of many immune cells such as helper / CD4 T lymphocytes and macrophages
HIV entry also requires a co-receptor on the host cell membrane, either CCR5 or CXCR4
HIV variants that bind to CXCR4 co-receptors infect helper/CD4 T lymphocytes
HIV variants that bind to CCR5 co-receptors infect macrophages.
binding of gp120 to receptors and co-receptors triggers a conformational change in the
viral envelope protein to expose a normally buried fusion peptide, gp41
gp41 mediates the fusion between the viral envelope and host cell surface membrane.
after fusion, the cone-shaped capsid is separated from the envelope
Describe penetration of HIV
once inside the host cell, the capsid proteins are digested by cellular enzymes, causing viral RNA and viral enzymes to be released. This process is called uncoating.
the cytoplasm, reverse transcription occurs, catalysed by viral reverse transcriptase.
viral (+) single-stranded RNA (ssRNA) is used as the template to synthesize a complementary single-stranded DNA (cDNA) by RNA-dependent DNA polymerase
ssRNA is then removed from the RNA-DNA hybrid by RNase
single-stranded cDNA is then used as a template to synthesize a linear double- stranded DNA by DNA-dependent DNA polymerase
Describe synthesis of HIV
When the host CD4 T lymphocytes are activated (stimulated during an immune response),
the integrated proviral DNA is transcribed by host DNA-dependent RNA polymerases
into viral mRNA molecules within the nucleus of the host cell.
o The viral (+)mRNA formed serves as:
i. new viral genome (to be incorporated into new viruses), which will then associate with
nucleocapsid proteins.
ii. template for translation (by host ribosomes on rough ER) into viral polyproteins (i.e.
a large protein that is cleaved into smaller proteins with different functions).
Note: HIV mRNA is polycistronic (i.e. one mRNA → many proteins) (Fig. 4.2f).
Fig. 4.2f: HIV mRNA is polycistronic.
Page 31 of 42o The viral polyproteins are then cleaved by HIV protease to form functional viral proteins
including:
➢ Structural proteins (e.g. capsid proteins, envelope glycoproteins, viral enzymes)
➢ Non-structural proteins
o Once the viral glycoproteins gp120 and gp41 are biochemically modified in the rER and
GA, they are transported by Golgi vesicles to the plasma membrane of the host cell.
When membrane of Golgi vesicle fuses with the host cell plasma membrane, these
glycoproteins are incorporated into the latter.
o The two linear (+) single-stranded RNA genome, two reverse transcriptase molecules,
HIV protease and integrase enzymes are encapsidated by the capsid proteins /
capsomeres.
Describe integration of HIV
newly formed double-stranded DNA migrates to the nucleus and integrates into the host genome
process is catalysed by the viral integrase
once integrate, viral double-stranded DNA becomes a permanent part of the host cell genome
o Once integrated, the viral DNA is known as a provirus.
DNA becomes a permanent part of the host cell genome.
once integrated, the viral DNA is known as a provirus.
host CD4 T lymphocytes can be activated to undergo cell division (“clonal expansion”).
enables the provirus to be replicated along with the host cellular DNA.
provirus can remain dormant (i.e. undetected by the immune system as it does not assemble into infectious viral particles) within a cell for a long period of time.
This is why AIDS patients typically show clinical symptoms only after a long latency period of 8-10 years.o
host CD4 T lymphocytes can be activated to undergo cell division (“clonal expansion”).
Describe release of HIV
newly assembled cone-shaped capsid and its contents will migrate to the host cell plasma membrane, where the viral glycoproteins gp120 and gp41 are incorporated, and bud off to form new viruses.
take away part of the host cell plasma membrane (containing gp120 and gp41) to form the viral envelopes.
budding itself does not necessarily kill the host cell, but eventually, the host cell will die due to depletion of energy and resources.
new viral particles are ready to infect other cells.
What is the affects the extent of damage of cell
extent of damage a virus causes depends partly on the ability of the infected tissue to regenerate by cell division
usually recover completely from colds because the epithelium of the respiratory tract can efficiently repair itself
however, damage inflicted by poliovirus to mature nerve cells is permanent because these cells do not divide and usually cannot be replaced
How do virus caused disease
direct cell damage and death may result from:
inhibition of normal host cell functions (DNA/RNA/protein synthesis) depletes the host cell of cellular materials (e.g. amino acids, nucleotides) essential for normal functions of the cell
may cause structural or functional defects in the infected host cells
some viral gene products are toxic, causing cell injury directly
indirect cell damage may result from:
attachment of viral proteins / glycoproteins (i.e. antigens) to the infected host cell surface membrane
result in an immune response as the infected host cell
may be recognised as foreign, and are thus destroyed by the body’s immune defences.
many of the temporary symptoms associated with viral infections, such as fever and aches, are a result of the body’s efforts at defending itself against infections.
integration of viral genome may result in certain types of human cancer (e.g. due to insertion of an oncogene or disruption of a tumour suppressor gene
How do vaccine help reduce virus infection
harmless versions of a specific virus (e.g. weakened virus, heat-killed virus, subunit of a virus, inactivated toxins produced by virus)
act as an antigen which stimulates an immune response (i.e. antibody production against the antigen)
constitute future defence, as when the body encounters the actual virus in future, the immune system will recognise and eliminate the virus quickly
Describe nucleotide mimics
inhibit viral DNA polymerases / RNA polymerases / reverse transcriptases
these prevent replication of viral genome and synthesis of viral proteins
Describe protein inhibitors
viral glycoprotein inhibitors → prevent entry (e.g. Enfuvirtide inhibits HIV gp41, hence prevents fusion/entry of virus) or release (e.g. Oseltamivir/Tamiflu inhibits neuraminidase of the influenza virus, hence prevents new viral particles from being released from the host cell)
viral ion channel inhibitor (e.g. Amantadine inhibits influenza M2 channel protein, hence prevents uncoating of the virus to release the viral genome)
viral enzyme inhibitor
Describe antigenic drift
a gradual process (i.e. occurs over a longer period of time)
involves minor mutations to the genes encoding the viral surface glycoproteins (e.g. influenza HA and NA which are involved in attachment and release of virus from host cells respectively).
one or more amino acids are changed, hence the shape of the glycoproteins (antigens) are slightly modified
makes it different from previous strains, hence it is necessary to reformulate the flu vaccine every year.
Describe antigenic shift
antigenic shift can only occur in viruses with segmented genomes, with genes found on each distinct segment (e.g. influenza virus).
it is a sudden process
involves a major change to the viral surface glycoproteins (e.g. influenza HA and NA which are involved in attachment and release of virus from host cells respectively)
not caused by a gene mutation but by a genetic exchange process, where entirely novel antigens are created
one than one strain of virus infects the same host cell (i.e. co-infection) - this is possible as some species (e.g. pig – the “mixing vessel”) are capable of being infected by different types of the virus. HA and NA from different strains of virus would be incorporated in the viral envelope of the new virion (surface antigens).
genetic reassortment occurs (i.e. segmented viral genome from different strains of virus are mixed together) during the process of viral replication, thus progeny virus has segments of genome from different strains of parental viruses
progeny virus will have a mixture of the surface antigens of the different strains of parental viruses, forming a new subtype.