Lecture 4: Antagonists in Functional Assays
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
23 Terms
Antagonism
A situation where adding two drugs into a system produces a lower response than would be expected in the drugs were added individually.
Simple Competitive Antagonists
When the antagonist binds to the same site as the agonist → results in a reduced response seen
Emax stays the same
EC50 increases ( curve shifts to the right) - as at increasing agonists concentrations this form of antagonism can be overcome → agonist can outcompete the antagonists
Functional Antagonism
Agonist binding results in a positive response; Agonist binding results in a negative response; responses from A and B cancel each other out
Non-Competitive Antagonists
Agonist and antagonist bind at separate binding site and no response occurs
Emax decreases
EC50 stays the same
Indirect Antagonism
Drug acting on a target causes the release of a transmitter that acts on receptor 1 to release a transmitter to activate receptor 2;
when an antagonist is bound, no response is seen as it blocks the actions of drug 1
Competitive Antagonist
Occurs when an antagonist binds to the same site as an agonist
It is an important mechanism from a therapeutic perspective, with many drugs acting at receptors through this mechanism
Gaddum Eqaution
Used to deal with competitive binding
Binding of a drug (D) is measured in the presence of a competing ligand, I.
Kd is the affinity of D,
Ki is the affinity of I
Bmax is the maximum binding the system can produce
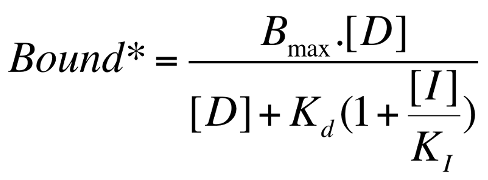
Gaddum Equation for Functional Measurements
Change B for binding into E for effect, and swap Ki for EC50
[D] is the concentration of the agonist,
[I] is the concentration of the competitive antagonist
KI is the equilbrium dissociation constant for the antagonist (i.e. the antagonist's Kd value).
![<ul><li><p><span>Change B for binding into E for effect, and swap Ki for EC50</span></p></li><li><p class="MsoNormal"><span> [D] is the concentration of the agonist,</span></p></li><li><p class="MsoNormal"><span>[I] is the concentration of the competitive antagonist</span></p></li><li><p class="MsoNormal"><span>K<sub>I</sub> is the equilbrium dissociation constant for the antagonist (i.e. the antagonist's K<sub>d</sub> value).</span></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/b39cac03-478d-448a-9c2b-3776aa23f4d4.png)
Measurements From Inhibition Assays
Use the Gaddum equation to study competition binding.
A fixed concentration of radiolabeled ligand is displaced by varying concentrations of a competing ligand.
This gives an IC₅₀: concentration of competitor that displaces 50% of the labelled ligand.
IC₅₀ depends on:
The concentration [D] and affinity (Kᴅ) of the radioligand.
The Cheng-Prusoff equation converts IC₅₀ to Kᵢ (true affinity of the competitor)
![<ul><li><p>Use the <strong>Gaddum equation</strong> to study <strong>competition binding</strong>.</p></li><li><p class="">A <strong>fixed concentration</strong> of radiolabeled ligand is displaced by <strong>varying concentrations</strong> of a <strong>competing ligand</strong>.</p></li><li><p class="">This gives an <strong>IC₅₀</strong>: concentration of competitor that displaces 50% of the labelled ligand.</p></li><li><p class=""><strong>IC₅₀ depends on</strong>:</p><ul><li><p class="">The <strong>concentration</strong> [D] and <strong>affinity (Kᴅ)</strong> of the radioligand.</p></li></ul></li><li><p class="">The <strong>Cheng-Prusoff equation</strong> converts IC₅₀ to <strong>Kᵢ</strong> (true affinity of the competitor)</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/7e9040d7-bac2-47ac-ada2-0870b0bc46e6.png)
Calculation of IC50
Normalise data to the control response (% of control).
Plot log(inhibitor) vs response and fit data using a 3-parameter inhibition equation (e.g., in Prism).
This yields an IC₅₀ (e.g., 2 nM).
Use a modified Cheng-Prusoff equation for functional assays:
Ki=IC501+[D]EC50Ki=1+EC50[D]IC50
Limitations of Method Used to Calculate IC50
Assumes competitive antagonism.
May not be valid with agonists showing cooperativity.
Assumes EC₅₀ ≈ Kᴅ, which may not always hold.
Difficulties in Applying Cheng Prusoff Equation
Assumes EC₅₀ = Kᴅ, which is rarely true in practice.
Assumes non-cooperative binding of the agonist.
Only valid for competitive antagonism—does not confirm that antagonism is competitive.
A more accurate method for functional data is Schild analysis, which specifically analyzes competitive antagonists.
Characeteristics of Competitive Antagonism
No change in Emax (maximum response remains the same).
Parallel rightward shift in the concentration-response curve.
Apparent increase in EC₅₀, now called EC₅₀* in the presence of an antagonist.
EC₅₀* increases in direct proportion to antagonist concentration [I].
This behaviour is described using the Gaddum equation.
![<ul><li><p><strong>No change in Emax</strong> (maximum response remains the same).</p></li><li><p class=""><strong>Parallel rightward shift</strong> in the concentration-response curve.</p></li><li><p class=""><strong>Apparent increase in EC₅₀</strong>, now called <strong>EC₅₀*</strong> in the presence of an antagonist.</p></li><li><p class=""><strong>EC₅₀*</strong> increases in <strong>direct proportion to antagonist concentration [I]</strong>.</p></li><li><p class="">This behaviour is described using the <strong>Gaddum equation</strong>.</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/b81f7b1d-5069-4bf9-84b5-fab1664b5f2d.png)
Schild equation
CR=1+[I]Ki
where CR = concentration ratio = EC50*/EC50
Derived by rearranging the Gaddum equation.
CR removes agonist-related variables (like EC₅₀ vs Kᴅ differences).
Makes analysis more reliable—agonist variability cancels out.
Developed by Heinz Otto Schild.
![<ul><li><p>CR=1+[I]Ki</p><p>where <strong>CR</strong> = concentration ratio = EC50*/EC50</p></li><li><p class="">Derived by <strong>rearranging the Gaddum equation</strong>.</p></li><li><p class=""><strong>CR removes agonist-related variables</strong> (like EC₅₀ vs Kᴅ differences).</p></li><li><p class="">Makes analysis more reliable—<strong>agonist variability cancels out</strong>.</p></li><li><p class="">Developed by <strong>Heinz Otto Schild</strong>.</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/fc93871c-26bd-41e7-a8fb-87f3234a1187.png)
Rearranged Schilds Equation
Log(CR-1) = log[I]-LogK1
CR=1+([I]/Ki); CR-1=[I]/Ki.
Now take logs: when taking logs, division becomes subtraction, so: log(CR-1) = log[i]-log Ki
This form of the equation allows for Log(CR-1) to be plotted against log {i{ and gives a linera graph with a slop of 1 and the y-axis intercept equal to -logkI
What Does Schilds Plot Reveal About Antagonism
Schild plot: log(CR − 1) vs log[Antagonist].
A straight line with slope = 1 suggests competitive antagonism.
If the line is not straight or the slope ≠ 1, the antagonism is not competitive.
Caution: Some non-competitive antagonists can also produce a slope ≈ 1 — additional tests may be needed.
How Does the Schild Plot Provide Information About Antagonist Potency
Y-axis intercept of the Schild plot = log(Kᵢ) → gives antagonist affinity.
From log(Kᵢ), calculate Kᵢ (dissociation constant).
X-axis intercept = when log(CR − 1) = 0, i.e. CR = 2, meaning the antagonist has doubled the apparent EC₅₀.
intercept gives a point of antagonist potency (where [I] causes a 2-fold shift in EC₅₀).
antagonist concentration at this point = measure of potency.
This value is expressed as a negative log concentration, called pA₂.
pA₂ = −log[antagonist] that gives CR = 2.
Plot must be linear for the interpretation to be valid.
![<ul><li><p class=""><strong>Y-axis intercept</strong> of the Schild plot = <strong>log(Kᵢ)</strong> → gives <strong>antagonist affinity</strong>.</p><ul><li><p class="">From log(Kᵢ), calculate <strong>Kᵢ</strong> (dissociation constant).</p></li></ul></li><li><p class=""><strong>X-axis intercept</strong> = when <strong>log(CR − 1) = 0</strong>, i.e. <strong>CR = 2</strong>, meaning the antagonist has <strong>doubled the apparent EC₅₀</strong>.</p><ul><li><p class="">intercept gives a point of <strong>antagonist potency</strong> (where [I] causes a 2-fold shift in EC₅₀).</p></li><li><p class=""> antagonist concentration at this point = <strong>measure of potency</strong>.</p></li><li><p class="">This value is expressed as a <strong>negative log concentration</strong>, called <strong>pA₂</strong>.</p></li><li><p class=""><strong>pA₂ = −log[antagonist]</strong> that gives <strong>CR = 2</strong>.</p></li></ul></li><li><p class="">Plot must be <strong>linear</strong> for the interpretation to be valid.</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/5f393f0d-5e86-4874-8e80-32a0e4ddf1f5.png)
pA2
"The negative logarithm to base 10 of the molar concentration of an antagonist that makes it necessary to double the concentration of the agonist needed to elicit the original submaximal response obtained in the absence of antagonist"
EC50
a "sub-maximal concentration" - pA2 in considered as the apparent EC50 value doubling
Relationship Between pA2 and Ki
For a perfect competitive antagonist, the log Ki value can be obtained either from the x or y intercepts:
x intercept (y = 0)
0 = log[I]-logKi
logKi = log [I]
y intercept (x=0)
log (CR-1) = -logKi
Therefore, if we have competitive antagonism, pA2 = -log Ki. Note, however, that this is not the definition of pA2.
F-Test
Compares two models:
Fixed slope = 1 (y = x + c)
Variable slope (y = mx + c)
Tests whether the more complex (variable slope) model fits significantly better.
If p < 0.05 → variable slope improves fit → not competitive antagonism.
If p > 0.05 → no significant improvement → use simpler model (slope = 1) → competitive antagonism likely.
Helps confirm the mechanism of antagonism
How Do 95% confidence intervals help in examining the slope in Schild plot analysis?
95% CI: Range of values where you can be 95% certain that the true slope lies'/ 5% chance that the true slope lies outside this range
If the 95% CI includes 1 (e.g. 0.7 to 1.1), competitive antagonism cannot be ruled out.
If the 95% CI does not include 1 (e.g. 0.7 to 0.9), competitive antagonism can be rejected with <5% chance of being true.
Helps determine if the data supports competitive antagonism or another mechanism.
Importance of pA2
High values indicates an antagonist of high potency and can indicate high specific of the antagonism
e.g., atropine blocks ACh receptors with low potency at histamine receptors
Similar values across different tissues suggest similar receptors in those tissues, better than comparing EC50 values, which can be affected by spare receptors.
Identical values for different agonists suggest they act on the same receptor, helping confirm receptor specificity.