BIS104 MT2
1/98
Earn XP
Description and Tags
Dinesh-Kumar Midterm 2 UC Davis
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
99 Terms
Lecture 9 04/21
Triton X-100
Solubilizes membranes while preserving protein structure
Replaces lipid bilayer with detergent micelles
Allows investigation of protein topology
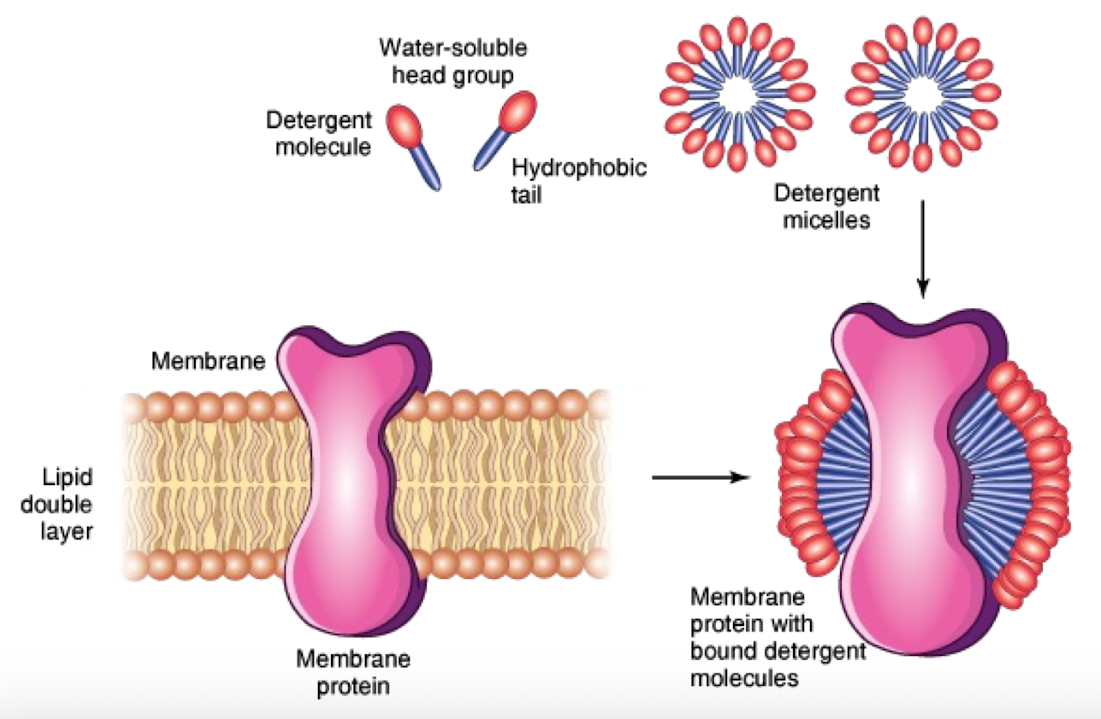
SDS Detergent
Stronger than triton X-100
Harsh, denatures proteins completely
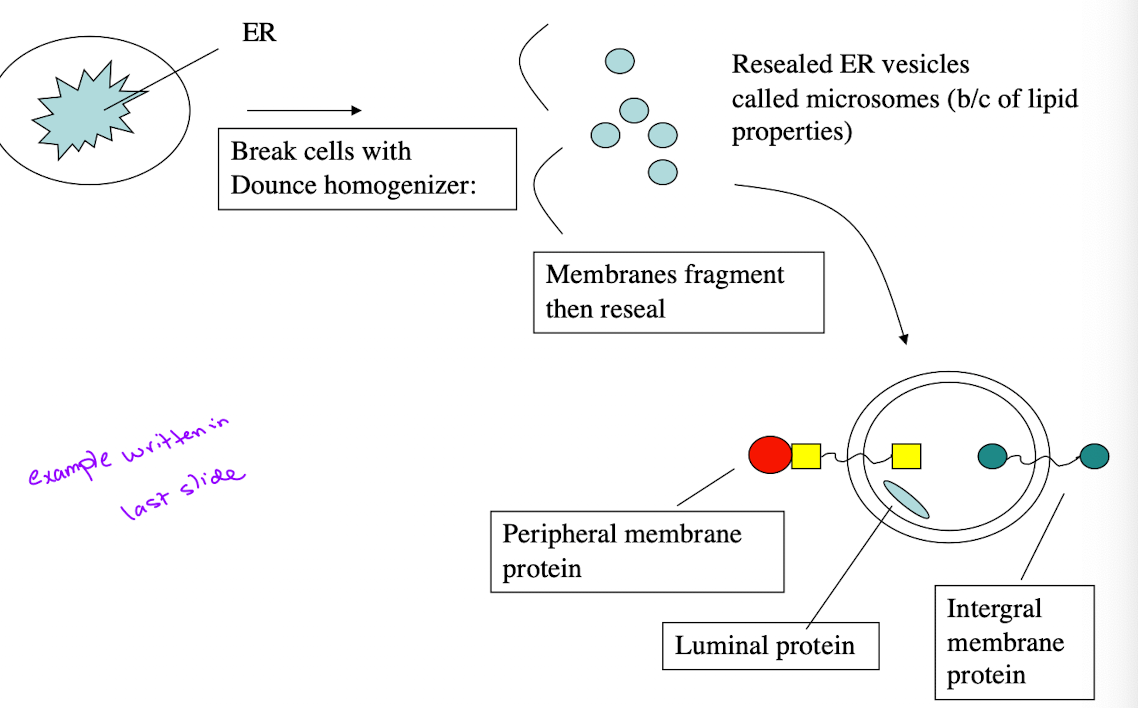
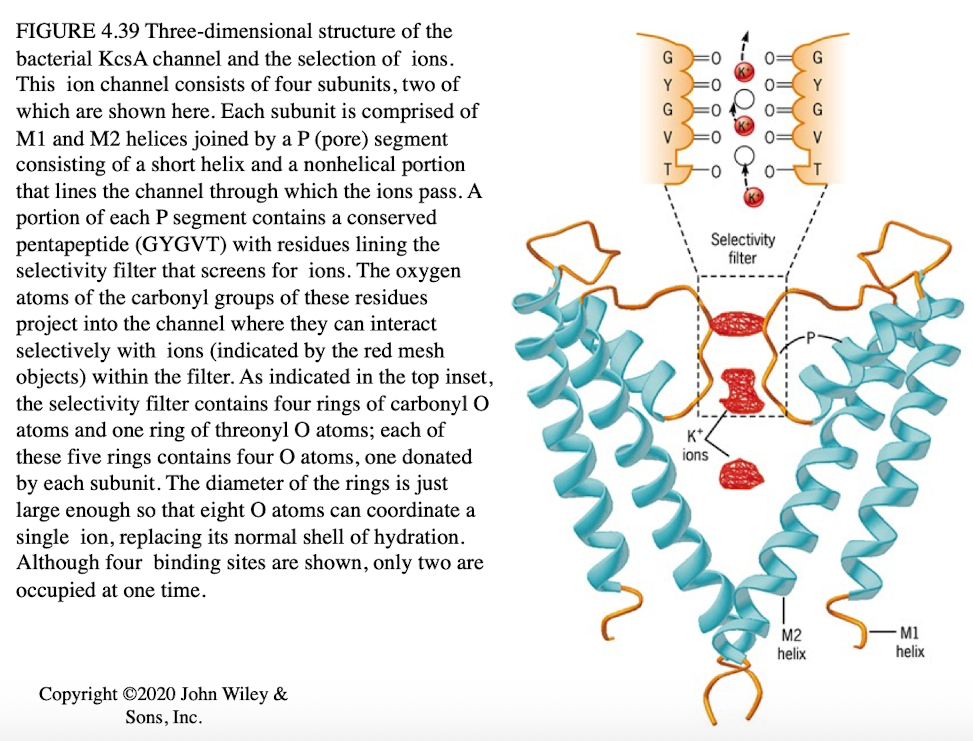
Trypsin Digestion Experiment
Purpose: Determine orientation of membrane proteins
Experimental Setup:
Homogenize cells/ER → microsomes
Add Triton X-100
Treat w/trypsin
Analyze SDS-PAGE
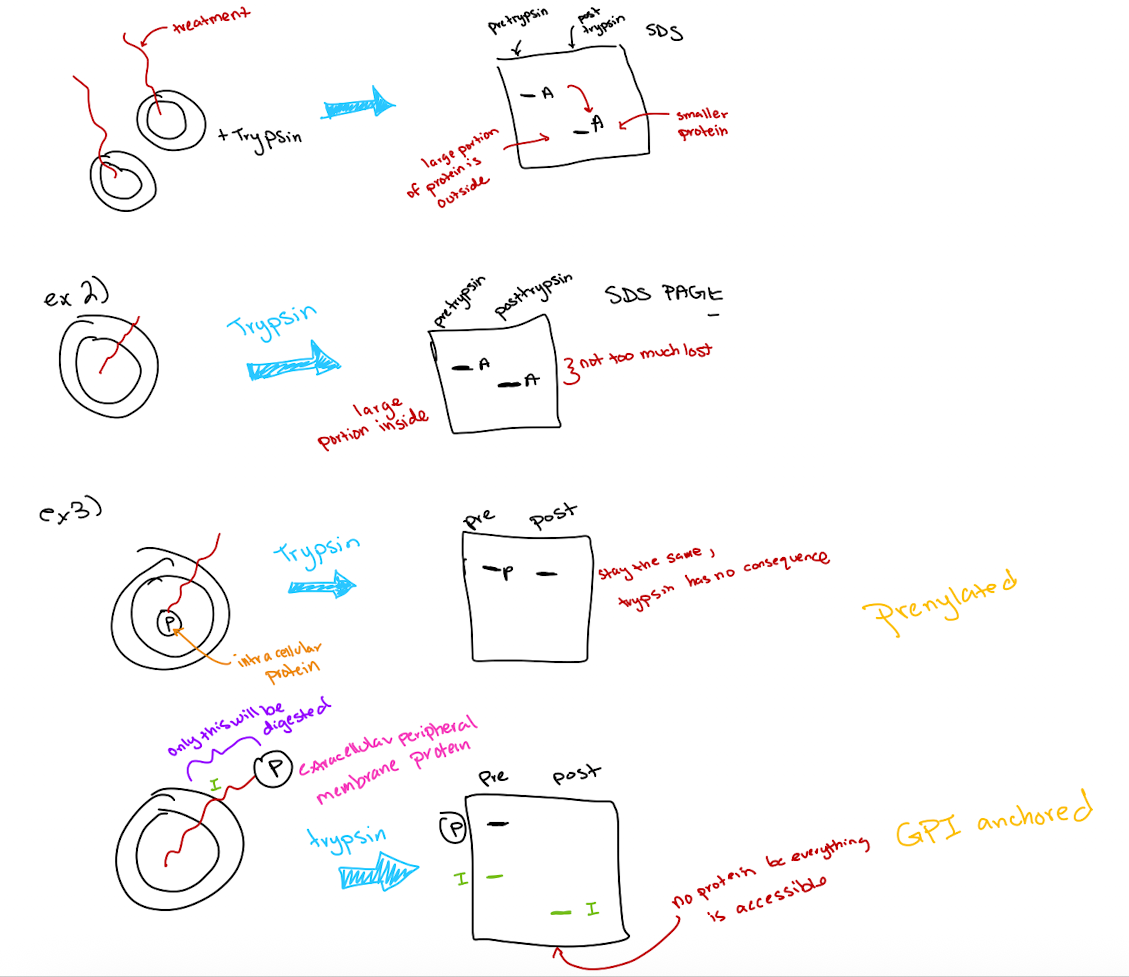
Trypsin Digestion Experiment Outcomes
Protein fragments indicate which portions were exposed
size change correlates with amount of protein digested
Large size reduction → large extracellular portion
little or no size change → mostly intracellular
Special cases
Peripheral proteins inside membrane: No size change
Peripheral proteins outside membrane: Completely digested
Selective permeability
Allows for separation and exchange of materials across the plasma membrane
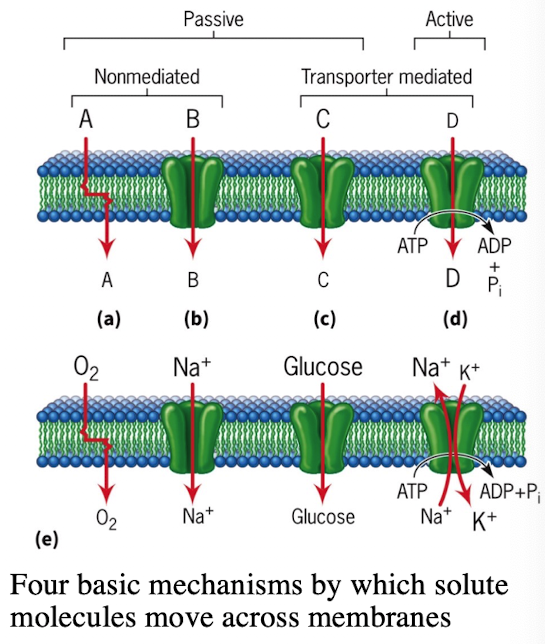
Types of Diffusion Across Membranes
Simple Diffusion: Small molecules (O2)
Passive Transport via Channels
Voltage-gated Na+ channels/leak channels
Voltage-gated K+ channels/leak channels
Rapid ion flow
Facilitated Diffusion via Transporters
Glucose (GLUT1, GLUT4)
Requires binding and conformational change
Active Transport via Pumps
Na+/K+ ATPase pump
Moves substances against concentration gradients
Requires ATP hydrolysis
Voltage-gated channels
conformational state depends on difference in ionic charge on 2 sides of the membrane
ligand-gated channels
conformational state depends on binding of specific molecule (ligand)
Mechanism-gated channels
conformational state depends on mechanical forces that are applied to the membrane
Potassium Ion Channel Specificity
Composed of 4 subunits
Pore region (P-domain) containing GYGVT peptide
Pore size matches K+ ions precisely
Sodium ions too small to interact effectively
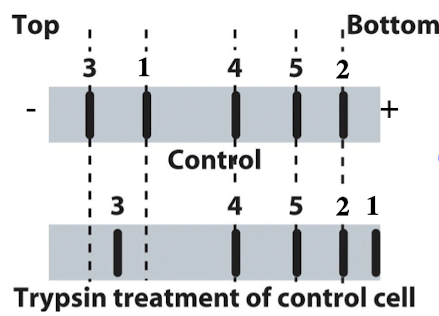
iClicker
You run proteins isolated from untreated control cells to trypsin treated cells on SDS-PAGE and transfer them to a membrane. You can detect 5 different proteins for which you have specific antibodies. What type of membrane protein is protein #5?
A. Transmembrane protein with equal domains both inside and outside the cell
B. Transmembrane protein with a large domain on the outside of the cell
C. Peripheral cytoplasmic protein
D. Peripheral extracellular protein
E. Transmembrane protein with large domain inside the cell
C. Peripheral cytoplasmic protein
Reasoning: No change after trypsin treatment, meaning no cleave and no access to inside of cell where cytoplasm would be.
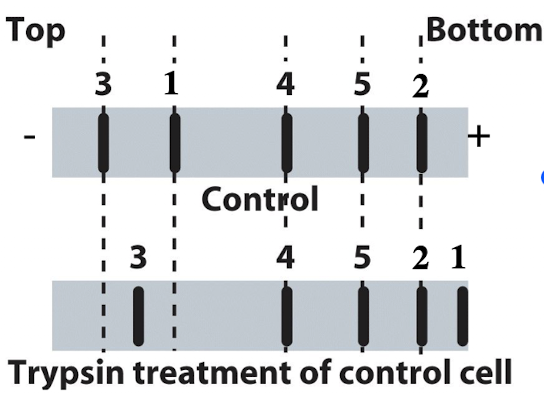
iClicker
You run proteins isolated from untreated control cells to trypsin treated cells on SDS-PAGE and transfer them to a membrane. You can detect 5 different proteins for which you have specific antibodies. What type of membrane protein is protein #1?
A. Transmembrane protein with equal domains both inside and outside the cell
B. Transmembrane protein with a large domain on the outside of the cell
C. Transmembrane protein with large domain inside the cell
D. Peripheral cytoplasmic protein
E. Peripheral extracellular protein
B. Transmembrane protein with a large domain on the outside of the cell
Reasoning: Protein becomes very small, making a lot of protein accessible to trypsin, making it outside of the cell
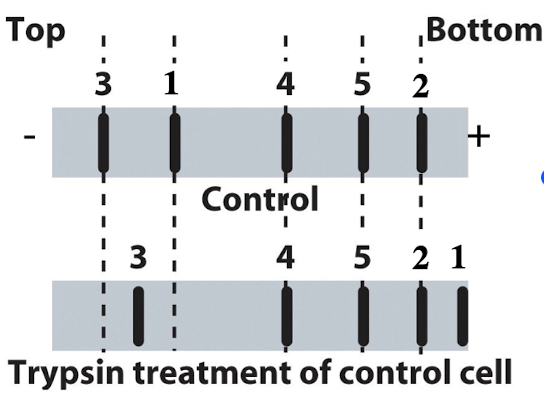
iClicker
You run proteins isolated from untreated control cells to trypsin treated cells on SDS-PAGE and transfer them to a membrane. You can detect 5 different proteins for which you have specific antibodies. What type of membrane protein is protein #3?
A. Transmembrane protein with a large domain on the outside
B. Transmembrane protein with large domain inside the cell
C. Transmembrane protein with domains both inside and outside the cell
D. Peripheral cytoplasmic protein
E. Peripheral extracellular protein
B. Transmembrane protein w/large domain inside the cell
Reasoning: Protein moved towards bottom when treated with trypsin
Lecture 10 04/23
Ion channels
passive transport, no energy required
rapid movement (millions of ions/sec)
Facilitated Diffusion by Transporters
slower than channels (hundreds to thousand ions/sec)
Requires selective binding and conformational change
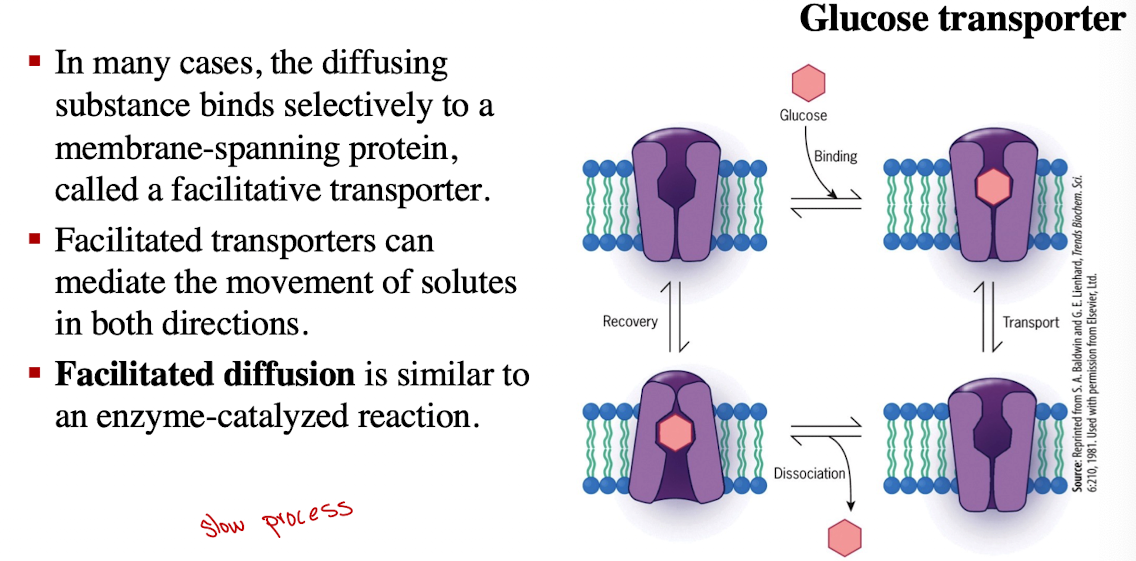
Facilitated Diffusion: Glucose Transporter
Glucose binds, transporter changes shape to release glucose inside cell
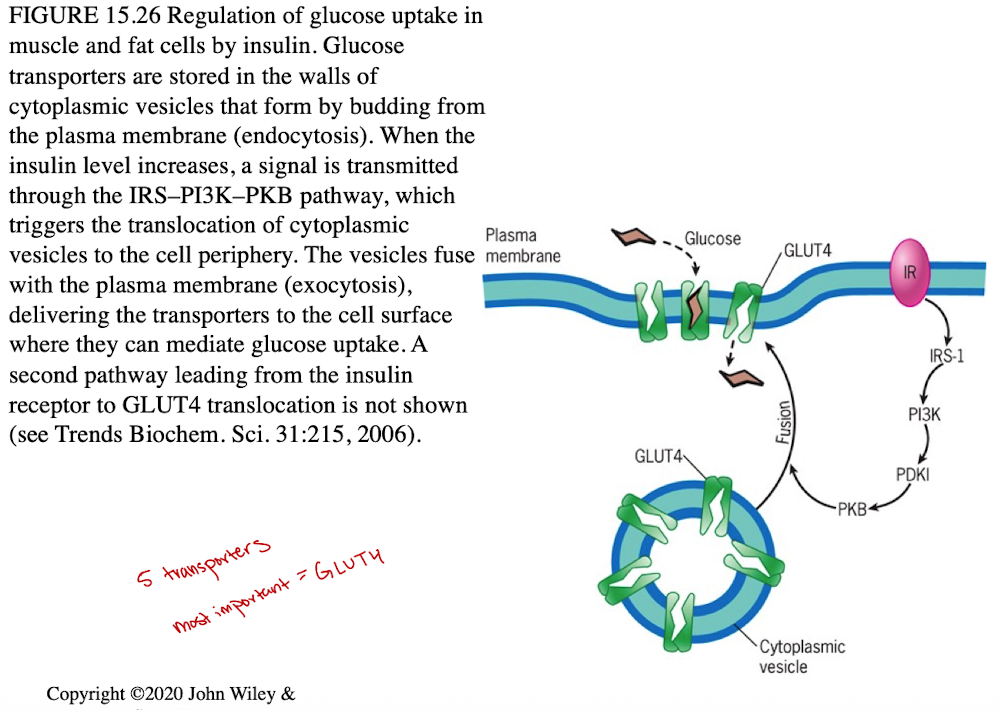
Insulin regulates GLUT4 (increase in blood glucose levels triggers secretion of insulin, stimulating the uptake of glucose)
Insulin triggers transporter to move to cell surface (membrane)
Active Transport
Maintains an imbalance of ions across the plasma membrane (moves molecules against conc. gradients)
Generate gradients
Requires coupled energy (ATP hydrolysis, absorbance of light, electron transport, flow of other substances down their gradients)
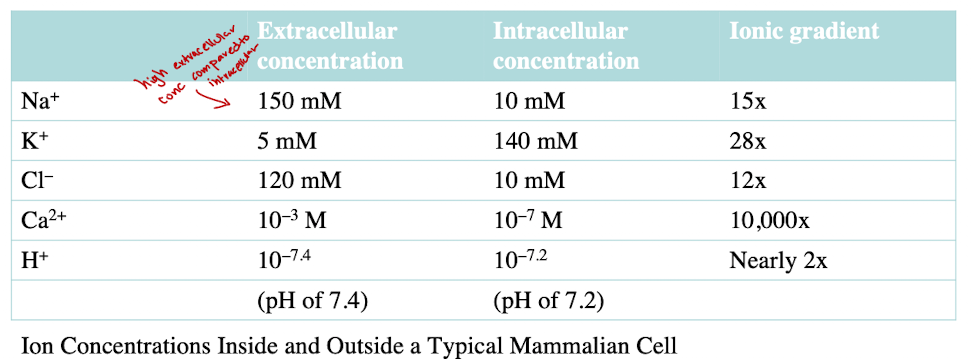
Concentration Gradients of Na+ and K+
Na+: high outside, low inside
K+: low outside, high inside
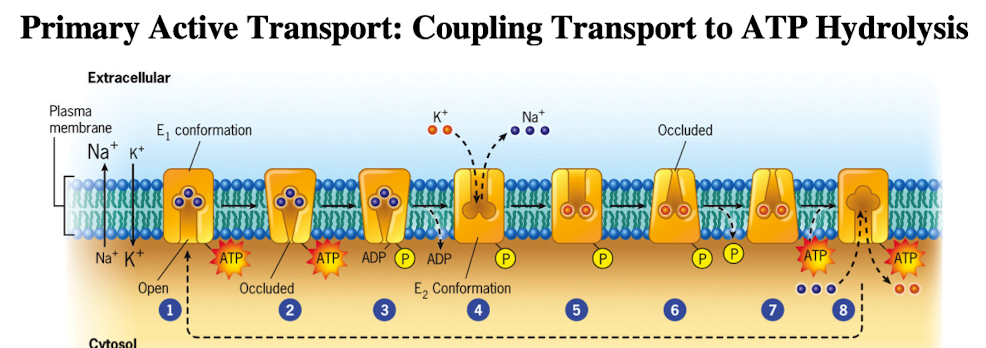
Primary Active Transport: Na+/K+ ATPase Pump
Requires K+ outside, Na+ inside and is inhibited by ouabain
3 Na+ out/ 2 K+ in per ATP hydrolyzed
Involves conformational changes, dephosphorylation and phosphorylation (ion affinity) bc it’s a P-type pump
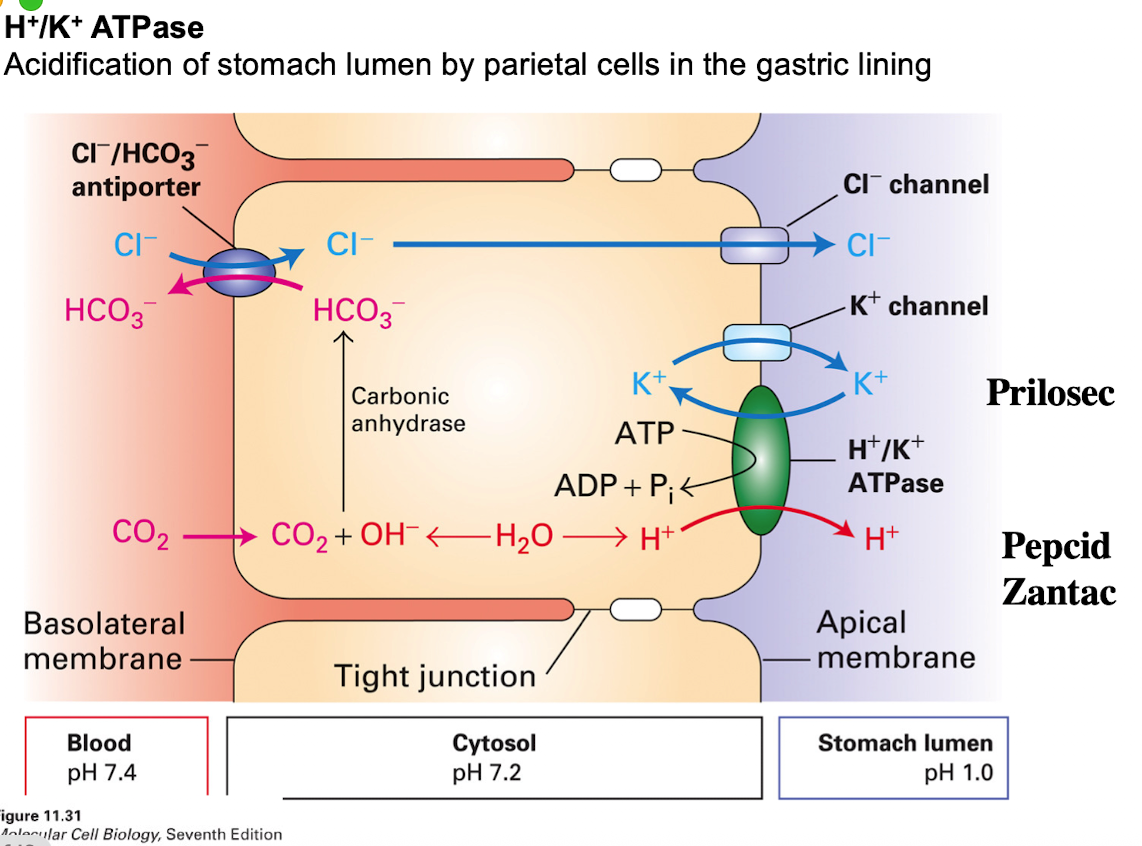
H+/K+ ATPase
maintains acidic pH in the stomach for digestion
Histamine signaling moves pumps to membrane
Treatments for acid reflux:
Antacids (Tums)
Histamine receptor blockers (Pepcid, Zantac)
Proton pump inhibitors (Prilosec)
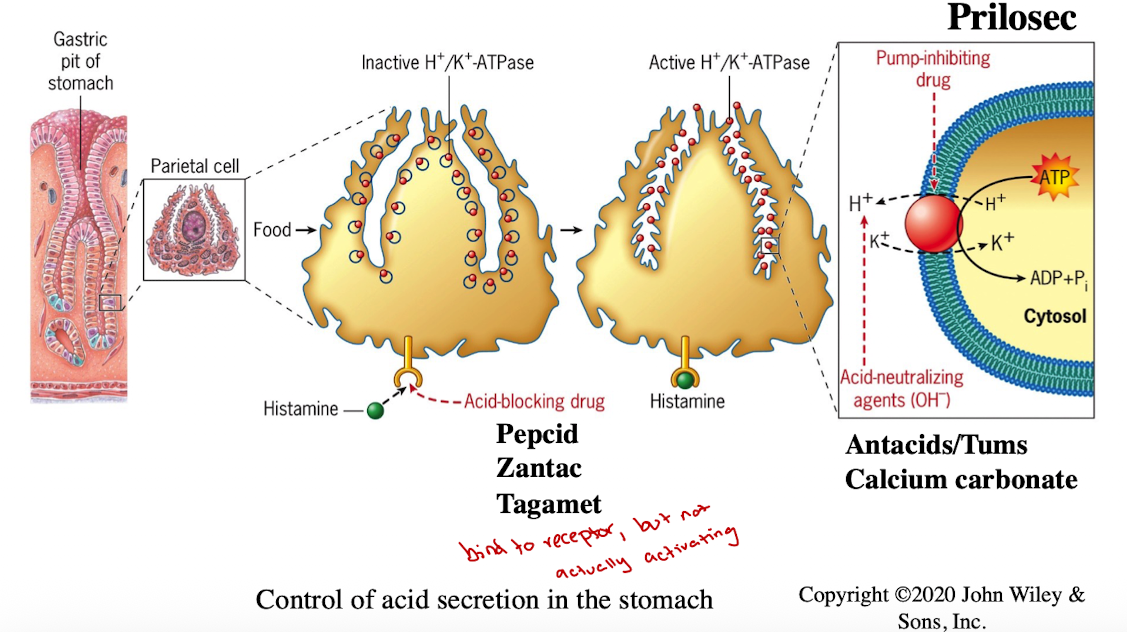
Acid Production in stomach
Channels, Pumps, and Transported all work together to help with CO2 uptake, ion exchange, K+ cycling)
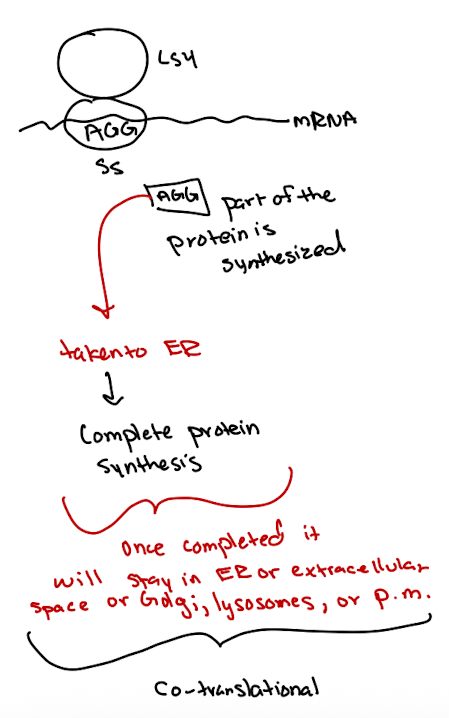
Protein Sorting Pathways: Co-translational
part of protein is synthesized
taken to ER to complete synthesis
Sorted to ER, Golgi, lysosomes, plasma membrane, or secreted into extracellular space
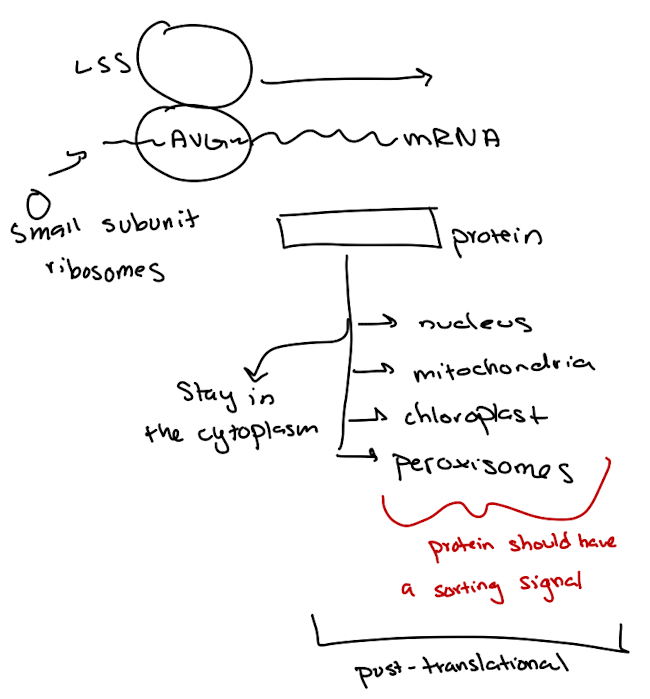
Protein Sorting Pathways: Post-translational
Fully synthesized proteins sorted based on targeting signals
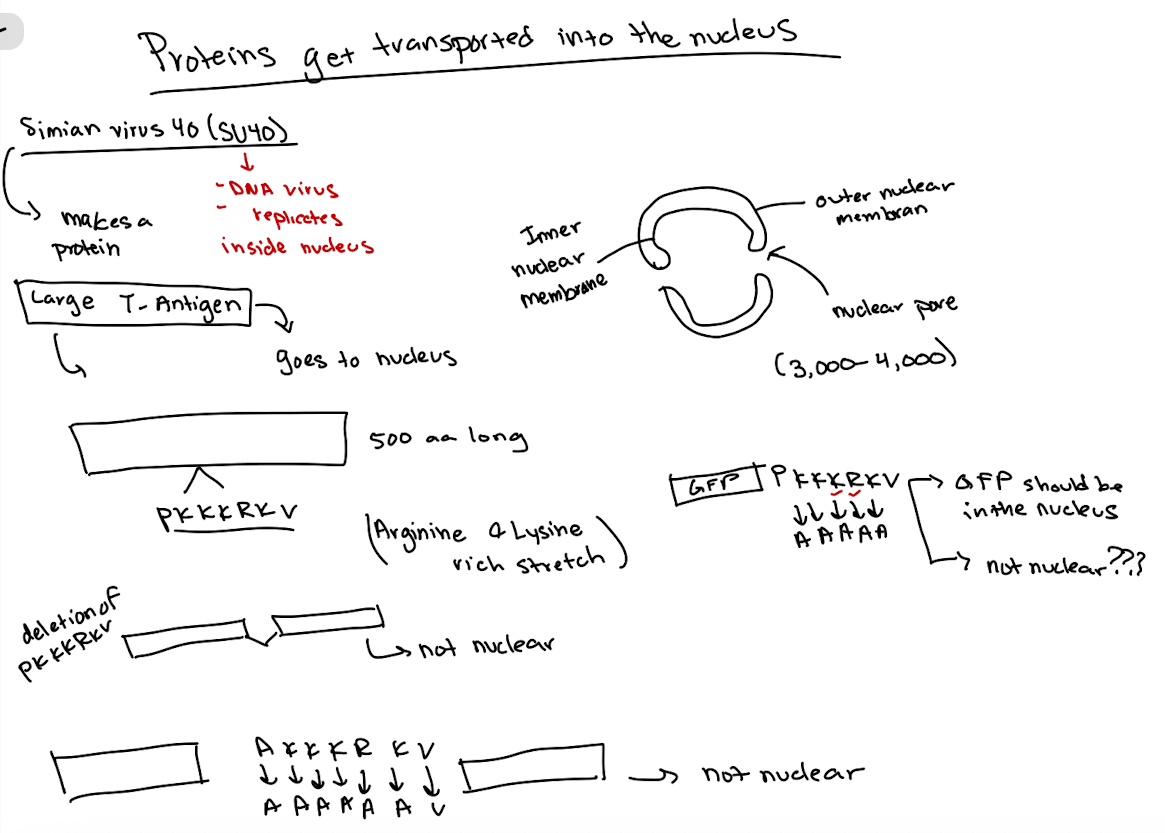
Nuclear Localization Signals
Identified using SV40 virus large T-antigen studies
Specific sequence (rich in lysine & arginine) directs nuclear entry
Presence of basic amino acid-rich signal suggests nuclear localization
Experimental approaches include:
deletion and point mutation experiments
fusion to GFP to test sufficiency
Lecture 11 04/28
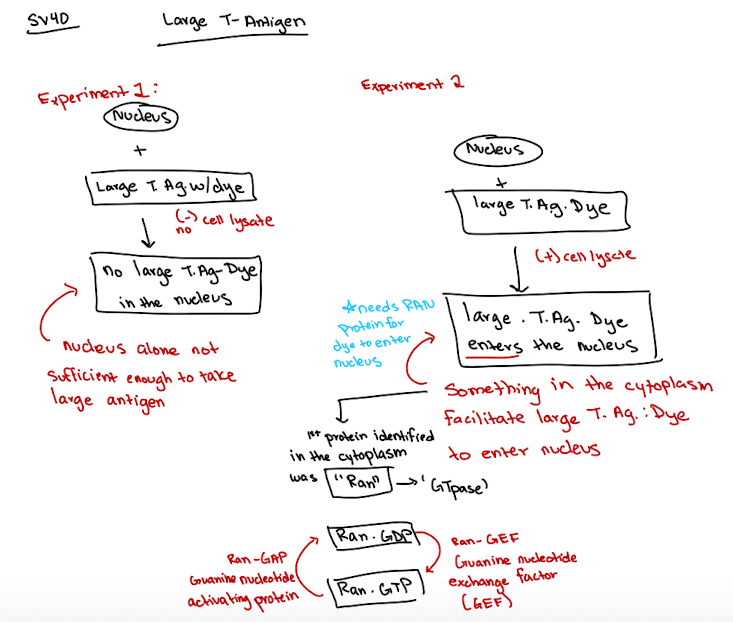
SV40 large T antigen experiment
nucleus alone is not sufficient for nuclear import and T antigen cannot enter (when it has no lysate)
Ran’s GTP/NDP state cycling is key for transport through the nuclear pore complex for large molecules
Ran is a GTPase function
Ran-GTP (active)
Ran-GDP (inactive)
Ran-GAP helps hydrolyze GTP → GDP
Ran-GEF helps Ran exchange GDP → GTP
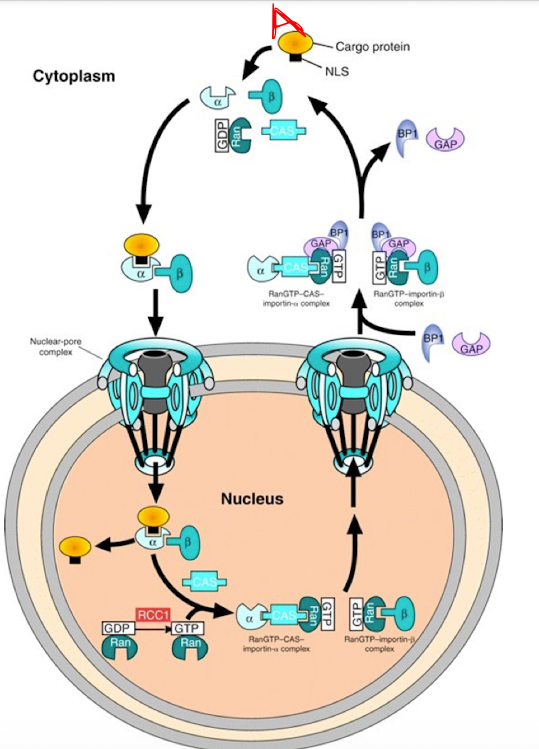
Nuclear Import Process
Cargo Recognition in Cytoplasm
Protein A (w/NLS) binds to Importin-α
Importin-α then recruits Importin-β → forms a trimeric complex (Protein A + α + β)
Transport Through Nuclear Pore
whole complex moves through the nuclear pore complex into the nucleus
Dissociation in Nucleus
Inside the nucleus, Ran-GTP binds to Importin-β, causing it to release
Then, CAS binds to Importin-α in the presence of Ran-GTP, helping release the cargo (Protein A) and exporting α
Recycling to Cytoplasm
α-CAS-Ran-GTP and β–Ran-GTP complexes diffuse back to the cytoplasm
Reset by Ran-GAP
In the cytoplasm, Ran-GAP converts Ran-GTP → Ran-GDP, which releases Importins
Ran-GDP goes back into the nucleus where RCC1 (Ran-GEF) regenerates Ran-GTP
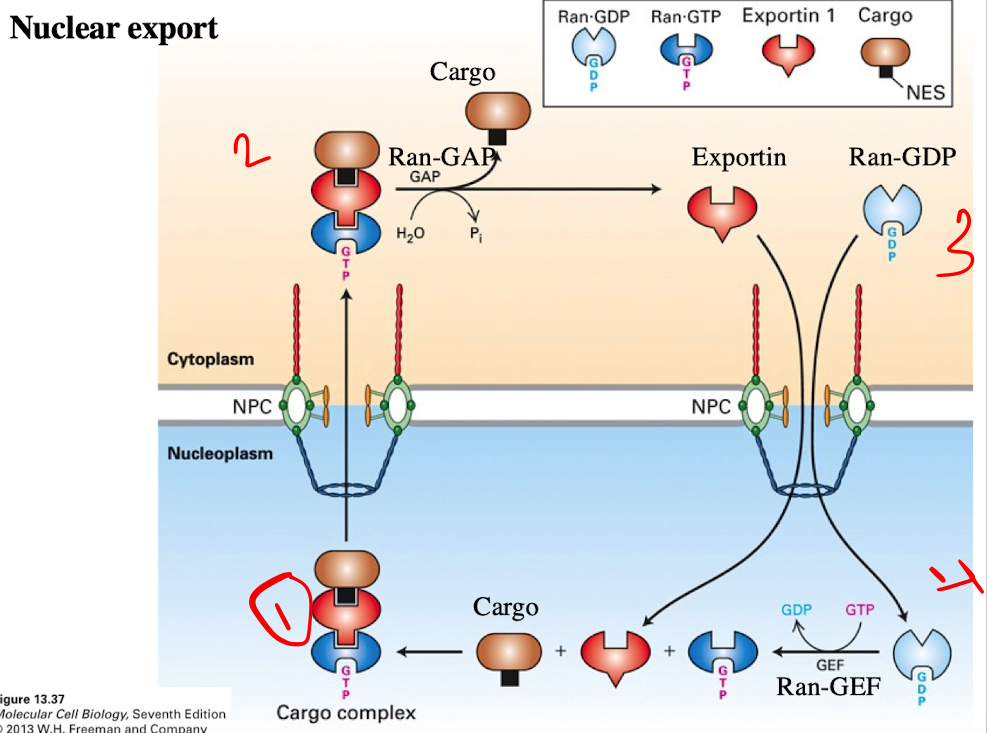
Nuclear Export Process
Cargo Binding (in the nucleus)
Exportin binds to the cargo protein with an NES
At the same time, Ran-GTP binds to exportin
Forms trimeric complex (Cargo + Export + Ran-GTP)
Translocation
Complex moves through the nuclear pore complex into the cytoplasm
Cargo Release
Ran-GAP in the cytoplasm converts Ran-GTP → Ran-GDP
This causes exporting to release the cargo protein
Exportin and Ran-GDP separate
Recycling
Exportin and Ran-GDP return to the nucleus
In the nucleus, RCC1 (Ran-GEF) converts Ran-GDP → Ran-GTP, resetting the system
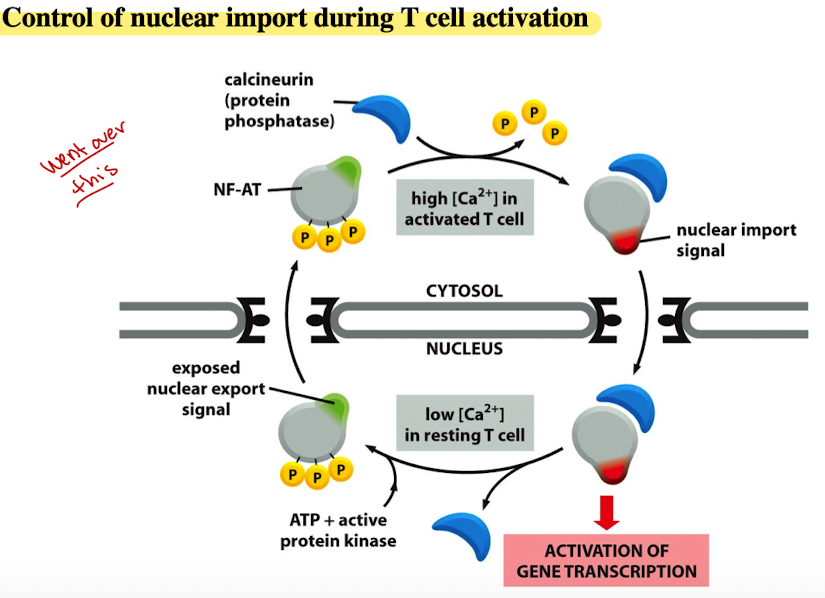
Control of nuclear import during T cell activation
Resting T Cell (low calcium)
In a resting T-cell, the conc. of calcium ions (Ca2+) in the cytosol is low
NF-AT is phosphorylated and resides in cytosol
ATP & active protein kinase maintain NF-AT in phosphorylated state, preventing entry into nucleus
Activated T cell (High Calcium)
When T cell is activated, Ca2+ conc. in cytosol increases
High Ca2+ levels activate calcineurin
Calcineurin dephosphorylates NF-AT, removing P groups
Nuclear Import
Dephosphorylated NF-AT exposes nuclear import signal
signal allows NF-AT enter the nucleus through nuclear pores
Gene Transcription
Once inside nucleus, NF-AT activates gene transcription
Leads to production of proteins necessary for T cell’s function in immune response
Nuclear Export and return to resting state
After T cell carried out function, Ca2+ levels decrease
NF-AT is re-phosphorylated by ATP and active protein kinase
Re-phosphorylation exposes nuclear export signal, causing NF-AT to exit nucleus and return to cytosol, ready for activation cycle
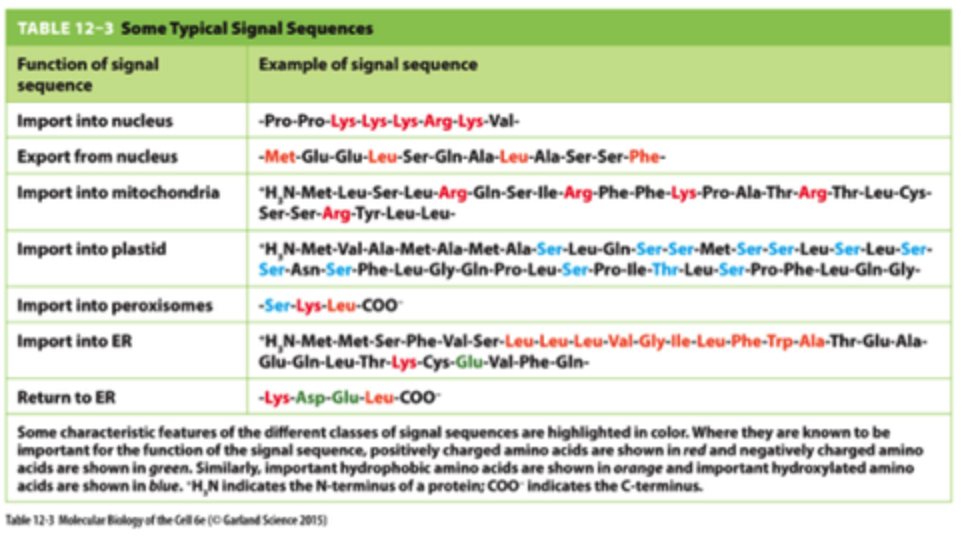
Signal Sequence Functions
lysine, arginine, proline, etc
iClicker
You are working with a protein, SUN1, which is typically
localized to the cytoplasm. If you add the PRRKKKRV
sequence to the beginning of the SUN1 protein, where would
you expect to find SUN1?
A. Cytoplasm
B. Mitochondria
C. Nucleus
D. ER
E. None of the above
C. Nucleus
Reasoning: Sequence is rich in arginine and lysine
iClicker
You are working with a protein, SUN1, which is typically localized to the cytoplasm. If you add the PRRKKKRV sequence to the middle of the SUN1 protein, where would you expect to find SUN1?
A. Cytoplasm
B. Mitochondria
C. ER
D. Nucleus
E. None of the above
D. Nucleus
Reasoning: post-translation, already made once put in
iClicker
In your research project, you have identified a protein that contains the 110VRRKKKRKP118 sequence. The fusion of this protein to YFP and observation using fluorescence microscopy indicate that this protein localizes to the nucleus.
Then, you mutated lysine residues at positions 113 to 115 to alanine, and the fusion of this mutant protein to RFP abolished nuclear localization.
What would you conclude from this experiment?
A. The sequence is sufficient to drive this protein into the nucleus
B. The sequence is necessary to drive this protein into the nucleus
C. Sequence is necessary and sufficient to drive this protein into the nucleus
D. None of the above
B. The sequence is necessary to drive this protein into the nucleus
Lecture 12 4/30
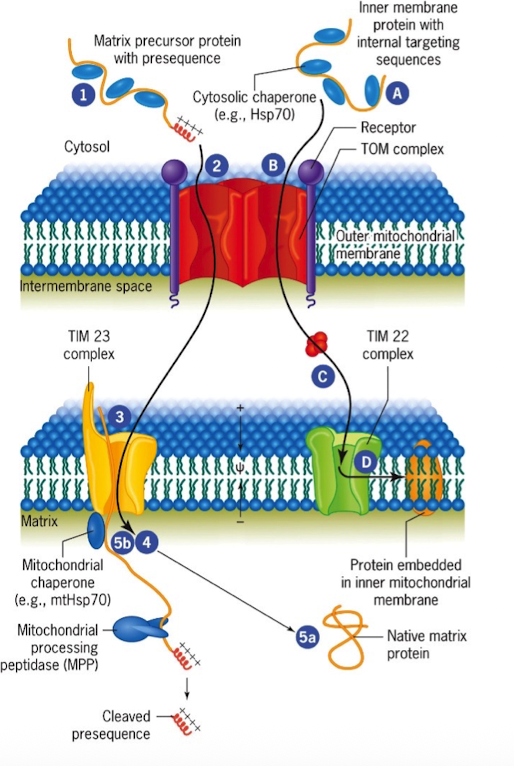
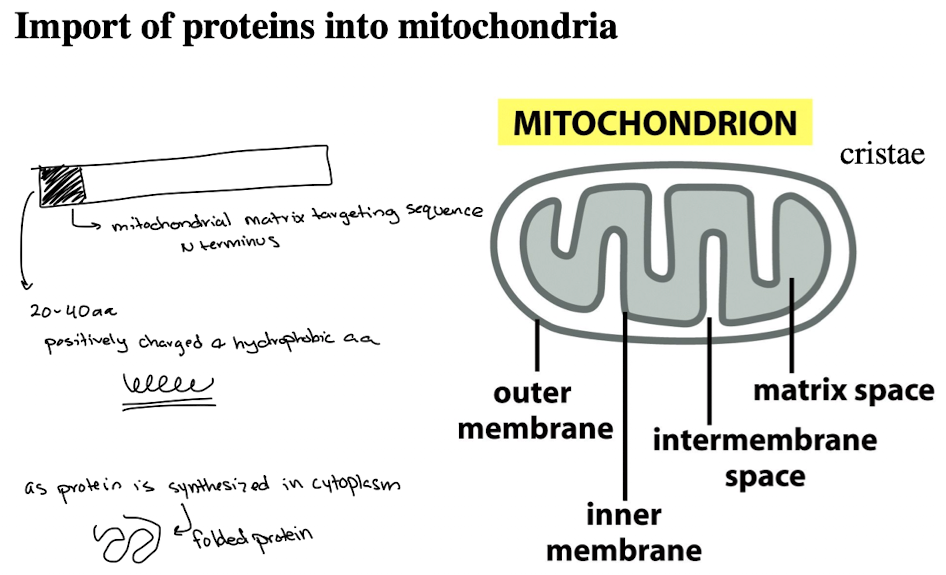
TOM Complex
Protein-import complex in the outer mitochondrial membrane that include receptor and channel (translocon of the outer membrane)

TIM Complex
Protein-import complex in the inner mitochondrial membrane (translocon of inner membrane)
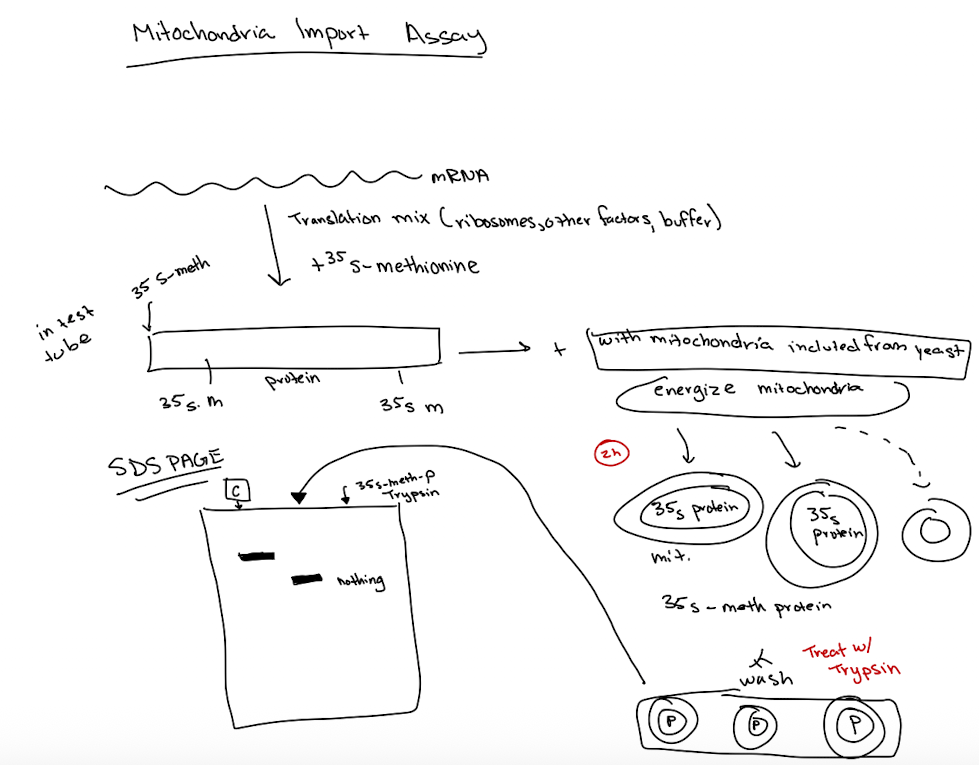
Mitochondria Import Assay
STEPS:
synthesize messenger RNA in vitro
Make protein in test tube using translation mic (ribosomes, buffers, other factors)
Radioactively label the protein with S35-methionine
Incubate labeled protein w/isolated mitochondria
Energize mitochondria to facilitate protein uptake
Wash mitochondria to remove unbound protein
treat w/trypsin to digest proteins outside mitochondria
separate proteins using SDS-PAGE
RESULTS: Smaller protein size in mitochondria due to cleavage of N-terminal terminal targeting sequence
N-terminal targeting sequence summary
The N-terminal targeting sequence is exposed early during protein synthesis.
Cytoplasmic chaperones bind to the protein to prevent complete folding and keep the N-terminal signal accessible.
The TOM complex recognizes this exposed N-terminal sequence.
The protein is unfolded as it is translocated across the mitochondrial membranes.
iClicker
You identified a new protein that contains a mitochondrial matrix targeting signal sequence. Where would you expect to find GFP if you fuse GFP to the N-terminus of the mitochondrial matrix signal sequence of the newly identified protein?
A. Mitochondria
B. Nucleus
C. Cytoplasm
D. ER
C. Cytoplasm
Reasoning: By placing GFP at the N-terminus, you're blocking the targeting signal from being first in line, which prevents recognition and import into mitochondria. As a result, the fusion protein fails to be imported into mitochondria and instead stays in the cytoplasm. Should be [Mitochondrial signal]-[GFP], not [GFP]-Mitochondria signal]
iClicker
You identified a new protein that contains a mitochondrial matrix targeting signal sequence. Where would you expect to find GFP if you put the mitochondrial matrix signal sequence of the newly identified protein in the middle of GFP?
A. Mitochondria
B. Nucleus
C. Cytoplasm
D. ER
C. Cytoplasm
Reasoning: Mitochondrial targeting signal sequence was placed in the middle of GFP. Needs to be placed at the N-terminus to be recognized, so protein will remain in the cytoplasm
iClicker
You identified a new protein that contains a mitochondrial matrix targeting signal sequence. Where would you expect to find GFP if you fuse GFP to the C-terminus of the mitochondrial matrix signal sequence of the newly identified protein?
A. Mitochondria
B. Nucleus
C. Cytoplasm
D. ER
A. Mitochondria
Reasoning: Targeting signal had to be correctly at the N-terminus to have been imported to the C-terminus of the mitochondrial matrix
TIM23
located in inner membrane
translocated proteins with N-terminal matrix-targeting signals
TIM22
located in inner membrane
inserts multipass inner membrane proteins
TOM
located in outer membrane
first entry point for all nuclear-encoded mitochondrial proteins
TIM9
located in intermembrane
chaperones hydrophobic proteins across the IMS to TIM22
TRP1
often used as a marker for gene disruption and plasmid construction bc mutants require tryptophan to grow (trp1 gene encodes synthesis of tryptophan)
iClicker
The yeast TRP1 gene encodes an enzyme that catalyzes the first step of tryptophan synthesis. TRP1 is a cytoplasmic protein. You are conducting an experiment in which you fuse the mitochondrial matrix signal sequence to the N-terminus of TRP1 (mitoMatrix-TRP1). Cells carrying mitoMatrix-TRP1 were grown in the absence of tryptophan. Most cells died, but some grew. Why did some cells grow in the absence of tryptophan?
A. Because of a mutation in the TRP1 gene
B. Because of a mutation in the Oxa1 gene
C. Because of a mutation in the Tim22 gene
D. Because of a mutation in the Tim23 gene
E. Because of a mutation in the Tim9 gene
D. Because of a mutation in the Tim23 gene
Reasoning: TIM23 disrupts import of mitoMatrix-TRP1 fusion into the mitochondrial matrix, so fusion protein stays in the cytoplasm, where TRP1 can function normally.
Lecture 13 5/2
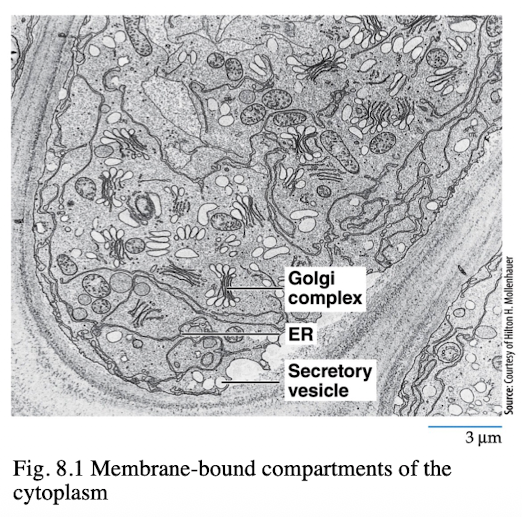
Endomembrane System
Formed by ER, Golgi complex, endoscopes, lysosomes, vacuoles that act as a coordinated unit
Distinct compartments bound by membrane barriers and contain specialized proteins for particular activities
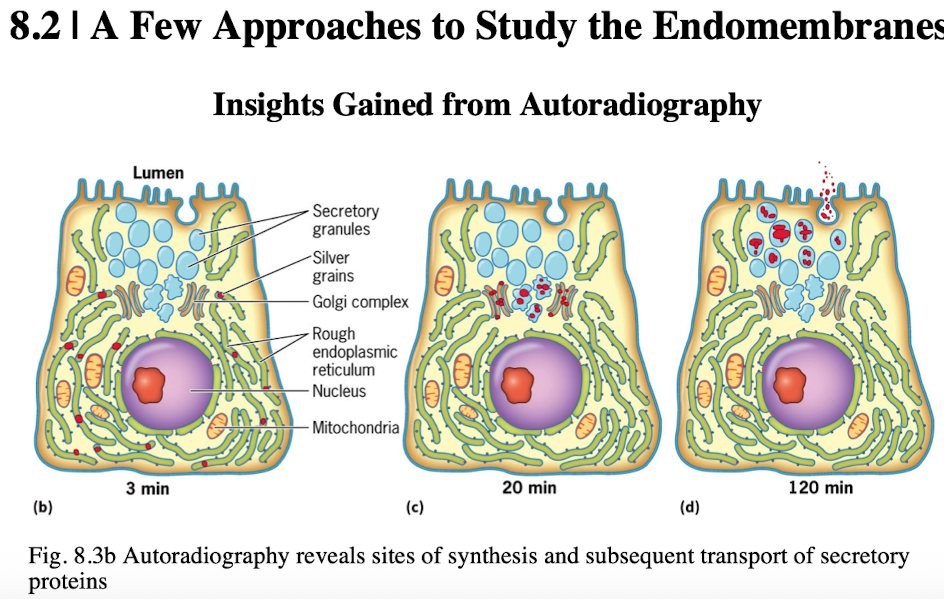
Autoradiography
visualizes biochemical processes by radioactively labeling molecules
Showed ER as site where secretory protein synthesis occurred
Palade and Jamieson incorporated radio labeled amino acids into pancreatic enzymes and were able to localize the cellular proteins
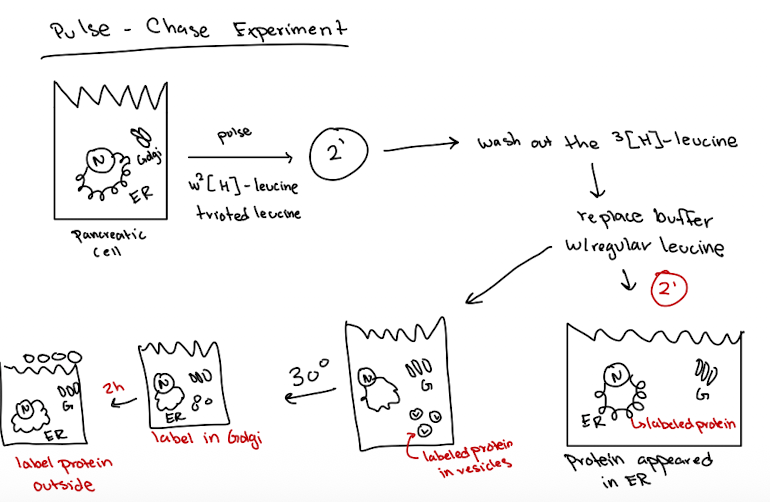
George Palade’s “Pulse-Chase” experiment
Pulse: Pancreatic cells were exposed to radioactive amino acid (S35-met) for a short time to label newly synthesized proteins
Chase: Cells were washed and incubated with non-radioactive amino acids to stop any new proteins from being labeled
Fixation and EM Imaging: At different time points during chased, cells were examined using electron microscopy and autoradiography
Results demonstrated secretory pathway of:
Rough ER → Golgi → Vesicles → Plasma Membrane (secretion)
Using Fluorescent Proteins in Endomembranes
GFP tagging allows microscope viewing of protein movement in living cells
Production and movement of viral proteins can be monitored
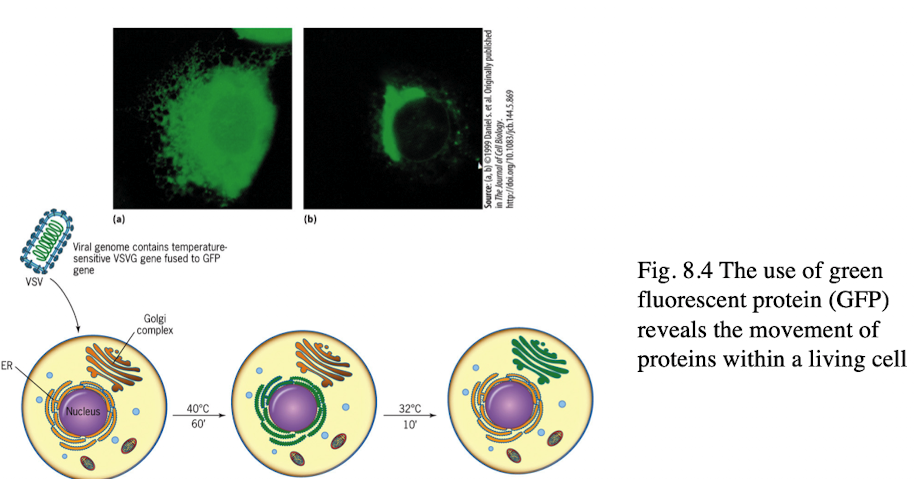
5 Clones of Sec Mutations
Sec A mutants: cytoplasm → ER
Sec B mutants: ER → vesicles
ER larger
Sec C mutants: Golgi → vesicles
excess vesicles from ER
Sec D mutants: vesicles → plasma membrane
protein gets stuck in Golgi
Sec E mutants: Plasma Membrane → secreted
protein gets stuck in vesicles btwn Golgi of pm
Rough ER (RER)
ribosomes bound to its cytosolic surface
flattened sacs (cisternae) connected to neighbors by helicoidal membranes
is continuous with the outer membrane of the nuclear envelope
starting point of biosynthetic pathway for secretory proteins
1/3 of proteins synthesized & released into ER lumen (co-translational translocation)
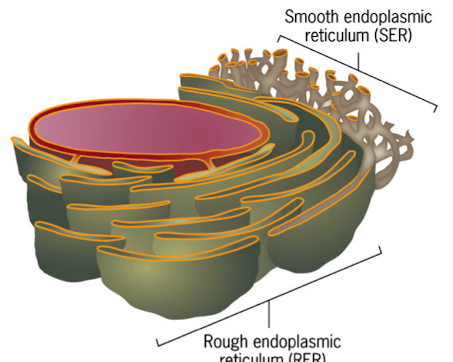
Smooth ER (SER)
lacks ribosomes
membranes are highly curved and tubular
is continuous w/the RER
calcium ion sequestration and regulated release
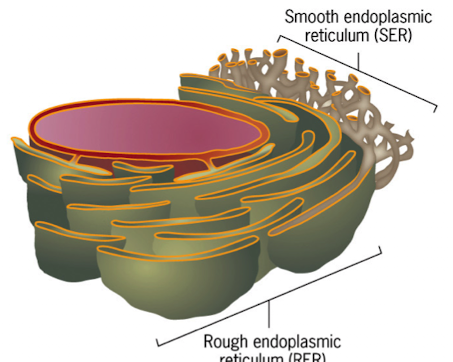
Gunter Blobel: Signal Hypothesis
Translation of secretory proteins w/o ER = proteins are longer and contain signal peptides
Translation w/ER present = shorter, processed proteins, confirming signal sequence was removed in the ER
Sec4
Function
Rab GTPase localized on secretory vesicles
Helps direct vesicles to pm, interacts w/effector proteins to promote tethering, facilitates vesicle docking & fusion w/pm in GTP-bound active form
After fusion, hydrolyzes GTP to GDP & recycled
Mutant
Vesicles cannot be properly targeted or docked to pm
post-Golgi secretory vesicles accumulate in cytoplasm
secretion blocked at final step after vesicle formation & Golgi exit
Sar1
Function
small GTPase that plays critical role in COPII vesicle formation, mediating ER to Golgi transport in secretory pathway
Activated by Sec12
Recruits sec23/24
Mutants of GDP-locked Sar1:
COPII vesicles cannot form
Proteins accumulate in ER, ER becomes enlarged, secretory pathway blocked
Mutants of GTP-locked Sar1
COPII coats do not disassemble
vesicles bud but don’t uncoat (fusion w/golgi blocked)
vesicles accumulate
Sec12
Function
Acts as GEF for Sar1 GTPase.
Catalyzes exchange of GDP for GTP on Sar1
GTP-bound Sar1 inserts into ER membrane and recruits COPII coat proteins (vesicle formation)
Mutant:
Sar1 remains GDP-bound and inactive.
COPII vesicle formation is blocked
Enlarged ER due to buildup of proteins
Sec17
Function
Disassembles SNARE complexes so they can be recycled
Works w/Sec18
Works after vesicle fusion
Mutant
SNARE complexes remain locked after fusion
Future vesicle fusion events are blocked (SNARE proteins not recycled)
Block in vesicle trafficking throughout secretory pathway
iClicker
A sec12 mutant yeast cell lack vesicles and has a larger ER as compared to wild-type. A sec17 mutant has an excess of vesicles and a small ER as compared to wild-type. What would you predict a sec12, sec17 double mutant would look like?
A. Normal number vesicles and a large ER
B. Transport from Golgi to secretory vesicles
C. Normal number of vesicles and a small ER
D. Too few vesicles and a large ER
E. Excess vesicles and a small ER
D. Too few vesicles and a large ER
Reason: If vesicles can’t be formed in first place (sec12), then the defect in vesicle fusion (sec17) becomes irrelevant.
iClicker
The Sec42 mutant has an excess of secreatory vesicles near the plasma membrane compared to wild-type. In the sec22 mutant, proteins that are supposed to be secreted accumulate in the Golgi. What would you predict a sec22, sec42 double Mutant would look like?
A. Enlarged ER and too few vesicles
B. Secreted proteins accumulate in Golgi
C. Excess secretory vesicles near the plasma membrane
D. None of the above
B. Secreted proteins accumulate in Golgi
Reason: cytoplasm → ER → vesicle → Golgi → vesicle → outside the cell. Proteins will accumulate in Golgi (sec22) before making it to vesicles near pm (sec42)
Lecture 14 5/5
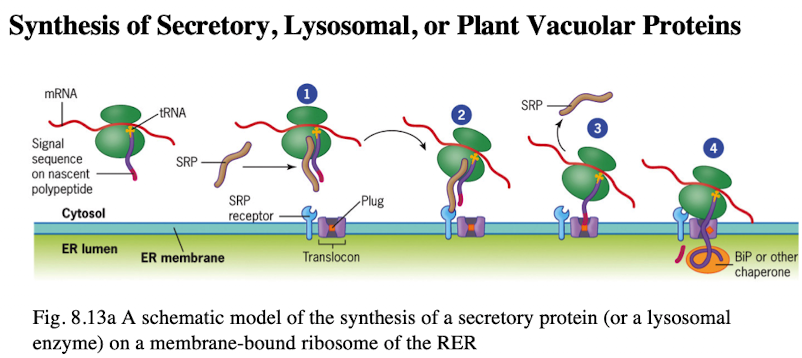
Synthesis of Secretory, Lysosomal, or plant vacuolar proteins
Co-translocation deposits protein into the ER lumen
Polypeptide signal sequence 6-15 hydrophobic amino acid resides, targeting pp to ER membrane
Signal sequence recognized by signal recognition particle (SRP)
SRP binds pp and ribosome, arresting synthesis
complex is recruited to ER thru interactions btwn SRP and SRP receptor
Ribosome goes to translocon. Once attached, signal sequence is recognized, pp is inserted into translocon channel

iClicker
Why is the band in lane 1 smaller than the band in lane 2?
A. The protein in lane 1 has a signal peptide
B. The protein in lane 2 has a signal peptide
C. The protein in lane 1 is a transmembrane protein
D. The protein in lane 2 is unfolded
B. The protein in lane 2 has a signal peptide

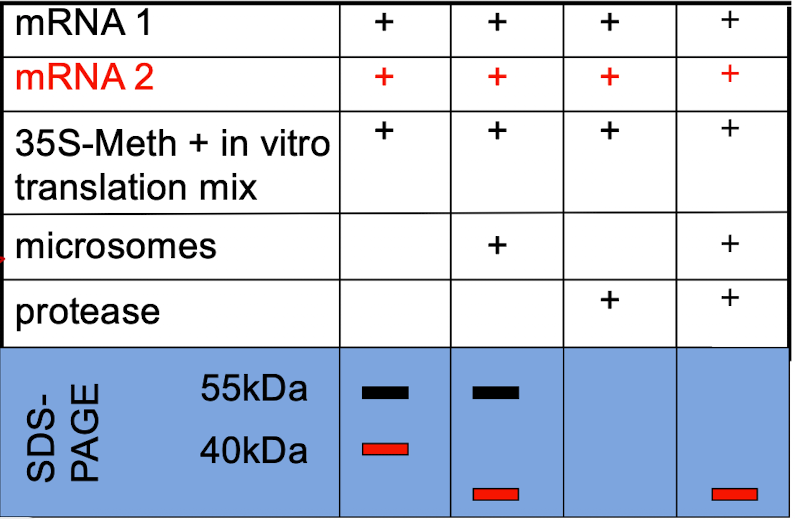
iClicker
Which protein would you suspect is secreted?
A. Protein 1 (black)
B. Protein 2 (red)
C. Both.
D. Neither one.
B. Protein 2 (red)
iClicker
If you add the nuclear localization sequence PKKKRKV to the beginning (N-terminus) of a secreted protein, where would you expect to find the protein?
A. The cytosol
B. The nucleus
C. The ER
D.Outside the cell
E. None of the above
D. Outside the cell
Reason: Secreted proteins already have an N-terminal ER signal sequence which directs the ribosome to the ER during translation. This signal overrides any downstream sequences like an NLS (PKKKRKV) and has no effect (following secretory pathway and ending up outside the cell)
iClicker
If you add the nuclear localization sequence PKKKRKV to the end (C-terminus) of an ER lumen protein, where would you expect to find the protein?
A. The cytosol
B. The nucleus
C. The ER
D.Outside the cell
E. None of the above
c. The ER
Reason: NLS must be in the cytoplasm to be recognized by importins that mediate nuclear import. Protein was already target to the ER lumen when NLS was added to C-terminus, so it stays bc no cytoplasm
iClicker
You have isolated a yeast strain with a mutation in the SRP. This mutant SRP can not bind nucleotides. Where would you predict to find a protein that is secreted?
A. Never made
B. Partially made, but still attached to ribosomes
C. In the cytoplasm
D. In the ER lumen
E. Secreted outside the cell
c. In the cytoplasm
Reason: SRP cannot bind, making it continue to translate in the cytoplasm and not be secreted.
Lecture 15 5/7
Type 1 membrane protein
N-terminus in lumen (outside cell)
single pass
signal peptide + stop-transfer sequence
C-terminus in cytoplasm
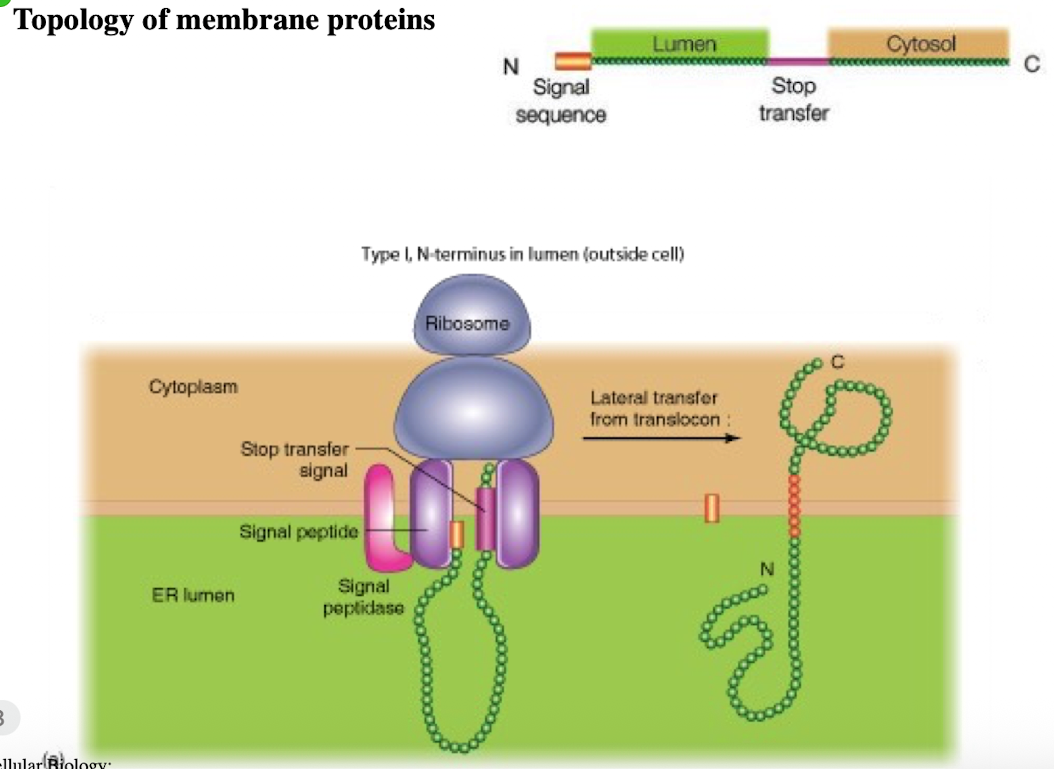
Type II membrane protein
N-terminus in cytoplasm (inside cell)
single pass
C-terminus in lumen
Internal signal-anchor sequence with (+) charged amino acids before internal sequence
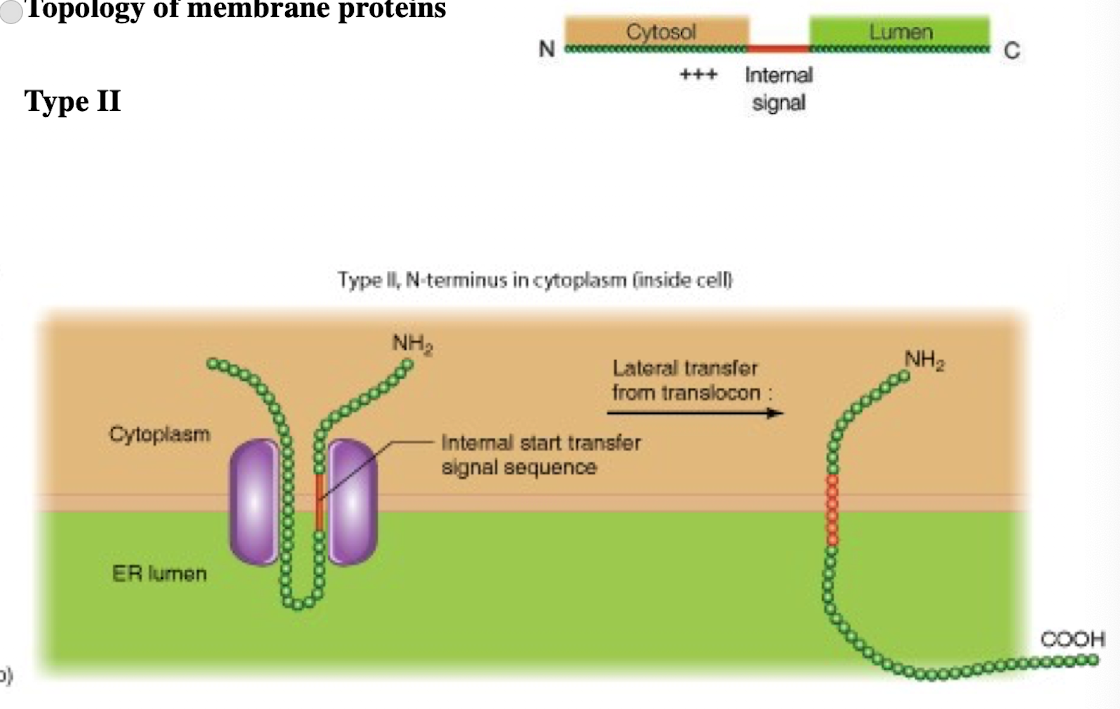
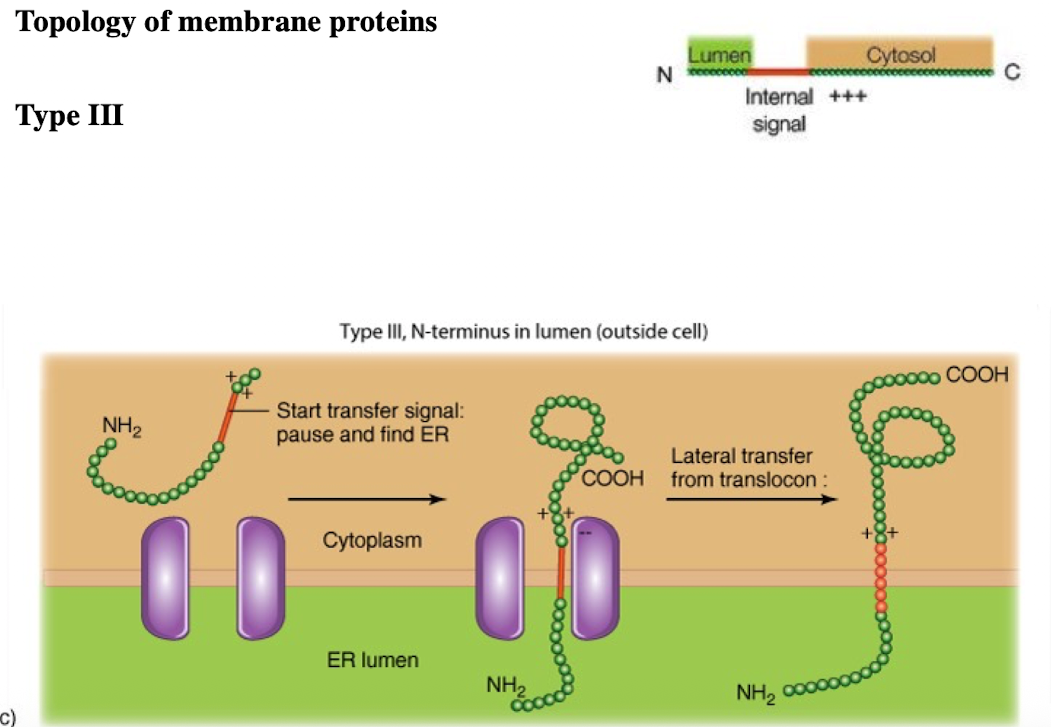
Type III membrane protein
N-terminus in ER lumen
single pass
C-terminus is in cytoplasm
internal signal anchor sequence with (+) charged amino acids after internal sequence
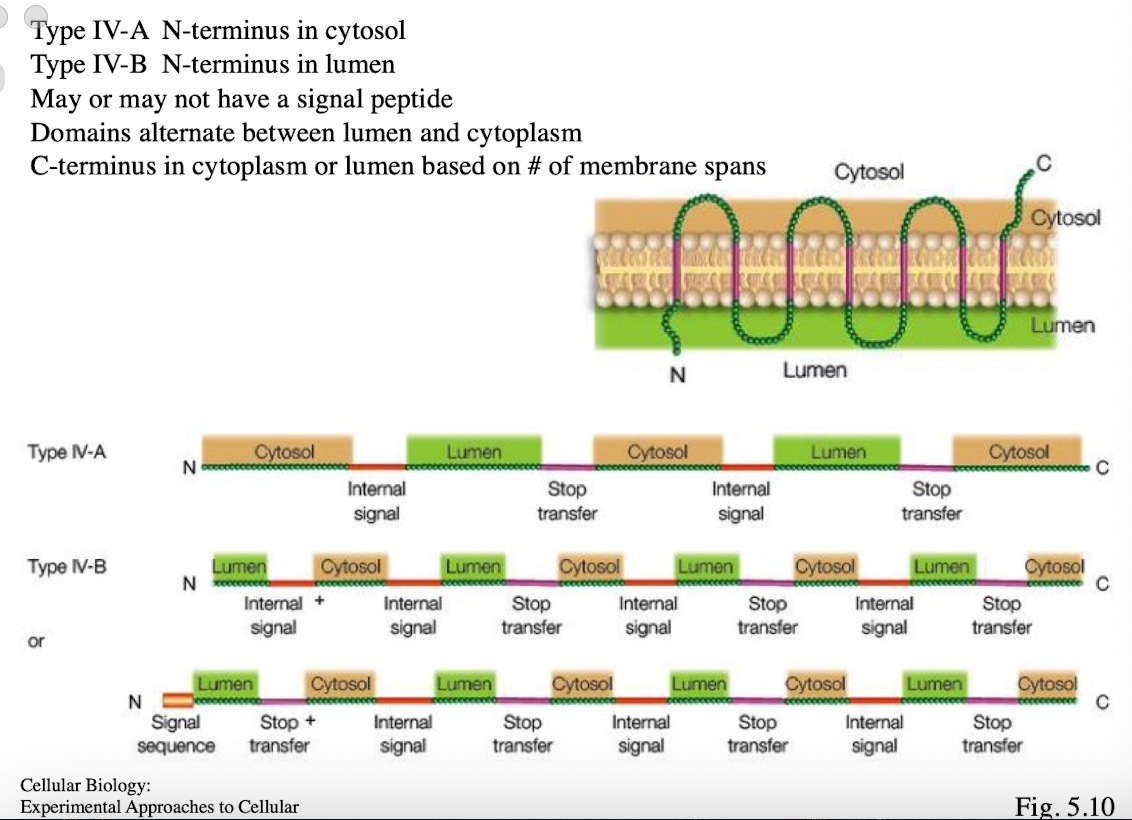
Type 4 membrane protein
Type IV-A: N-terminus in cytosol
Type IV-B: N-terminus in lumen
multi-pass
Multiple internal start and stop sequences
may or may not have signal peptide
domains alternate btwn lumen and cytoplasm
C-terminus in cytoplasm or lumen based on # of membrane spans
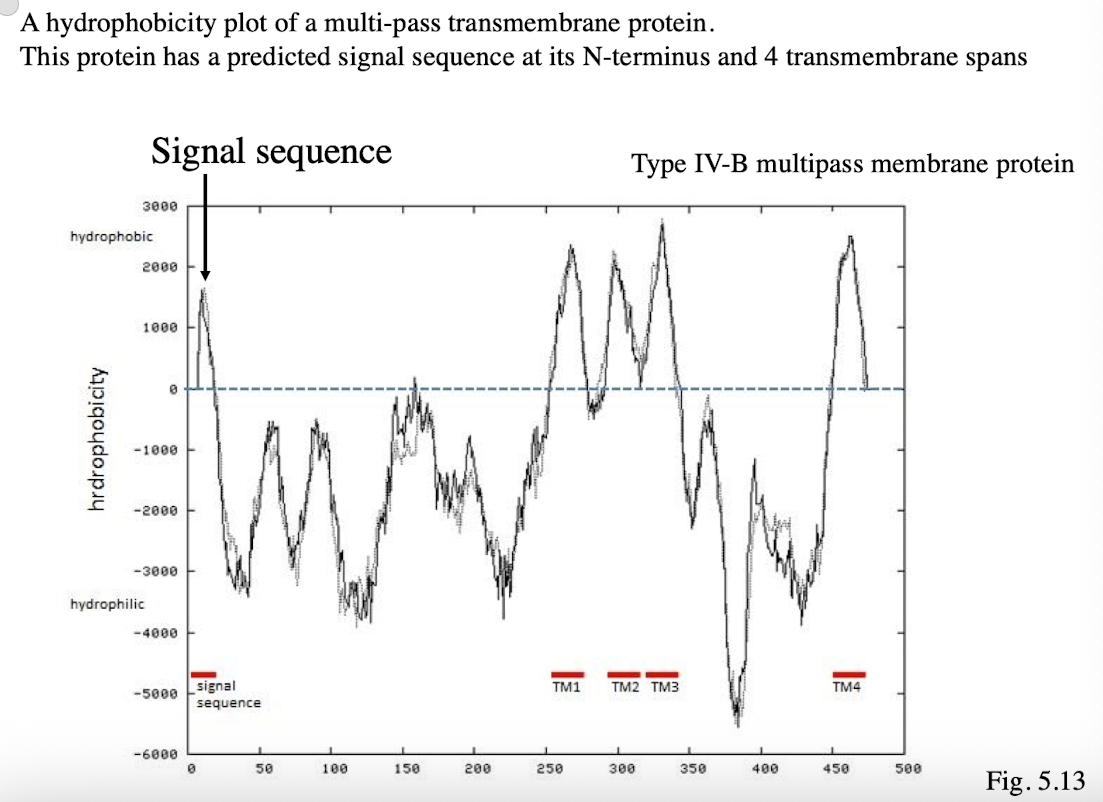
Hydrophobicity Plot of Type IV-B
Protein has predicted signal sequence at its N-terminus (first peak) and 4 transmembrane spans (peaks after signal sequence)
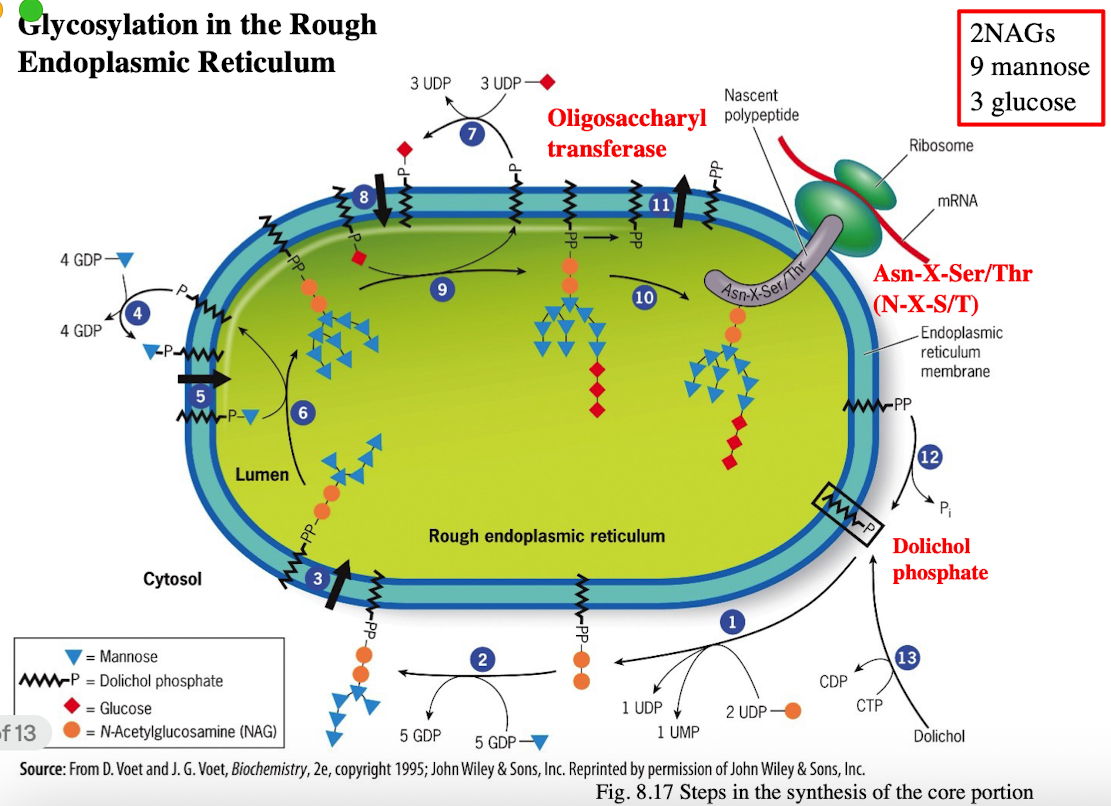
Glycosylation in RER
Attachment of an Oligosaccharide (N-linked Glycosylation)
Catalyzed by oligosaccharyltransferase
Glucose and mannose residues are trimmed to prepare for further folding and processing
Glycan helps proper folding by interacting with chaperones like calnexin and calreticulin
Misfolded proteins will be glucose tagged, mannose deficient and degraded by proteasome (ER)
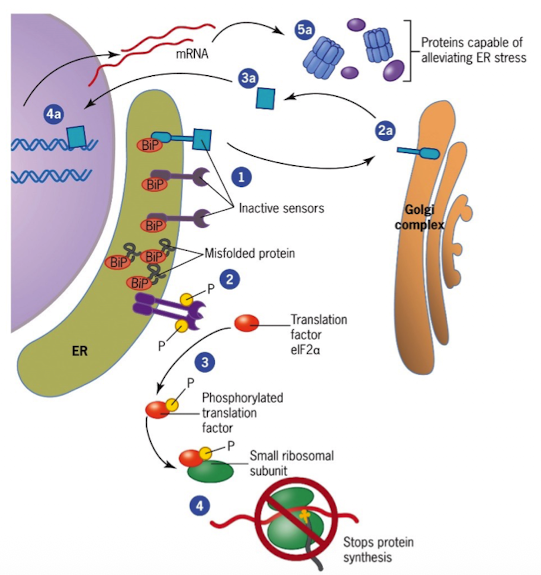
Mechanisms that Ensure Destruction of Misfolded Proteins (UPR)
Accumulation of misfolded proteins triggers the unfolded protein response (UPR)
Sensors in the ER are kept inactive by the chaperone BiP (BiP binds to unfolded proteins and sensors)
When misfolded proteins accumulate, BiP is incapable of inhibiting sensors
activated sensors send signals to trigger proteins in destruction of misfolded proteins
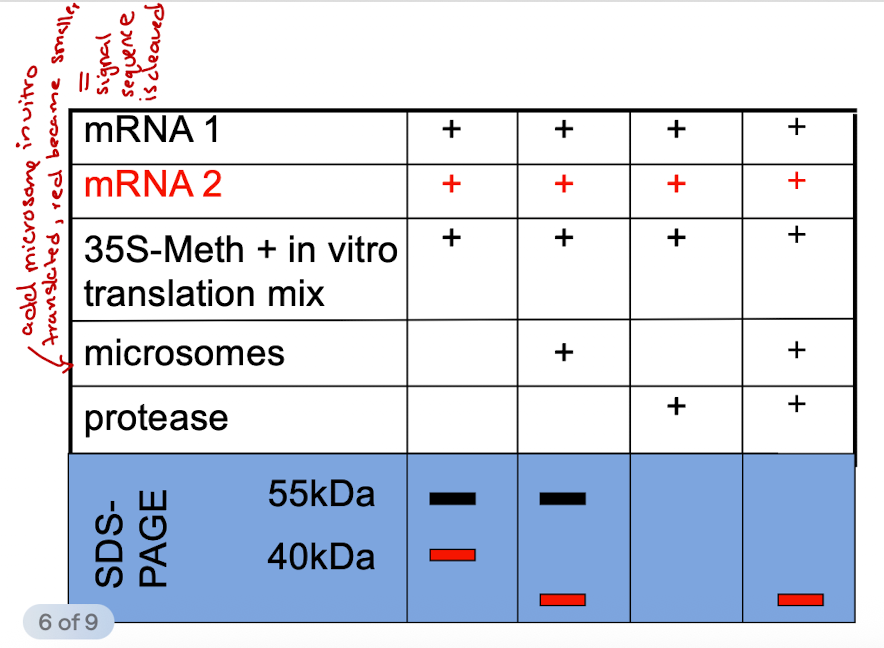
iClicker
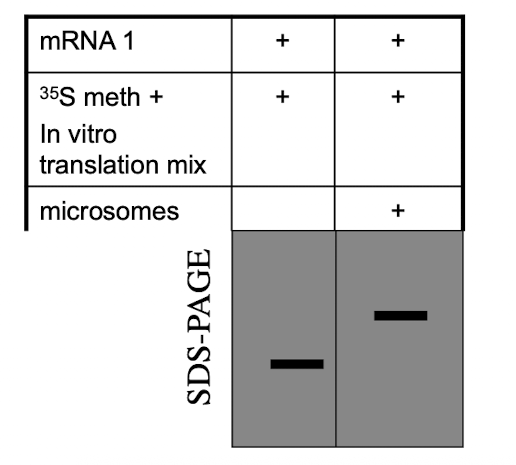
Hypothesize why is the band in lane 1 smaller than the band in lane 2?
A. The protein in lane 2 is unfolded
B. The band in lane 1 has had its signal peptide cleaved
C. The protein encoded for by mRNA 1 is a multipass membrane protein.
D. The band in lane 2 has a signal peptide
E. Sugars were added to the protein in lane 2
E. Sugars were added to the protein in lane 2
Reason: Protein 2 has microsomes, which act like ER and allow glycosylation to happen. Sugars are added to the protein, making it larger so it runs slower on SDS-PAGE.
Lecture 16 05/09
How are materials carried between compartments
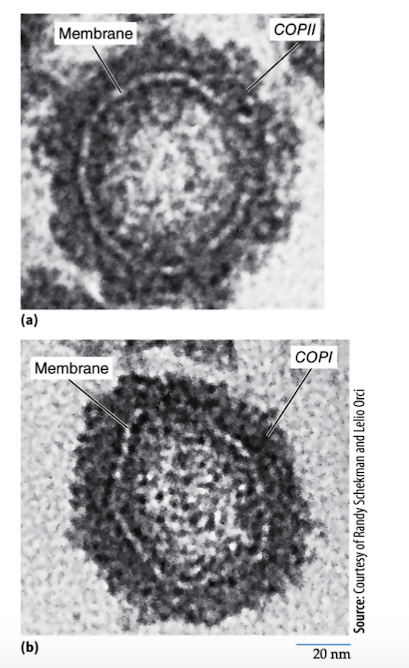
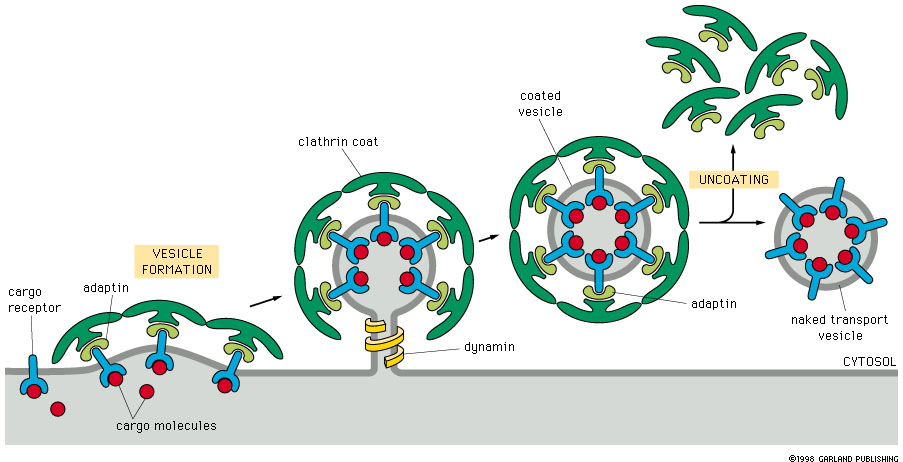
using coated vesicles
Protein coats 2 functions
Cause the membrane to curve and form a vesicle
Select the components to be carried by vesicle
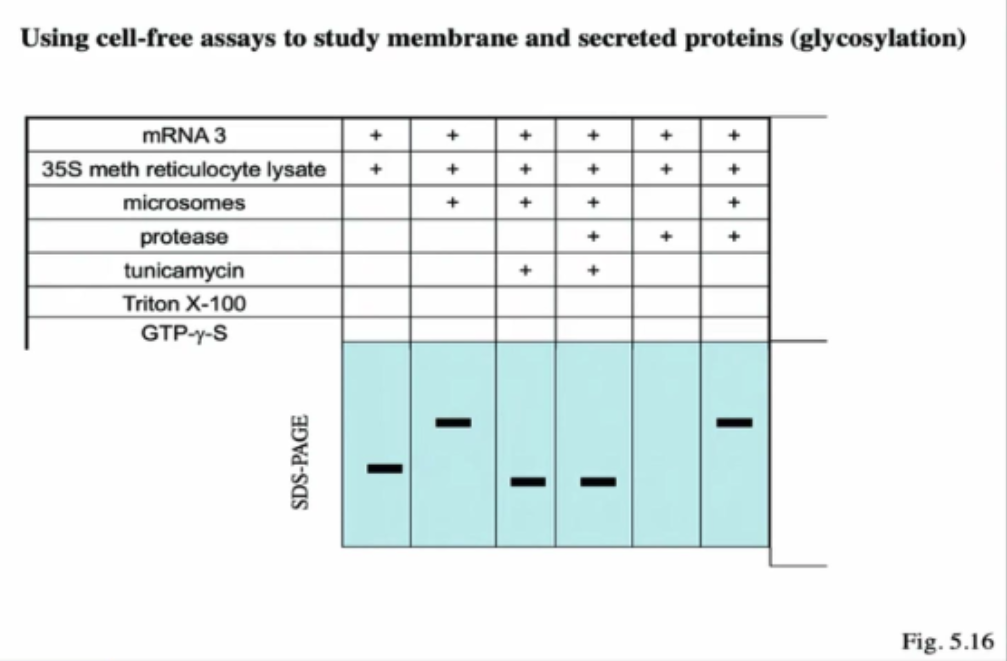
Using cell-free assays to study membrane and secreted proteins (glycosylation)
Lane 1
Only mRNA and the lysate
Lane 2
mRNA + lysate + microsomes → L2 has a larger size than L1 bc L2 undergone glycosylation
Lane 3
mRNA + lysate + microsomes + tunicamysin → Lane 3 is smaller than L2 and L1 bc tunicamycin will block glycosylation (aka sugars aren't added, and its smaller than Lane 1 bc the signal sequence was cleaved inside microsome)
Lane 4
mRNA + lysate + microsomes + the protease + tunicamycin → L4 has the same band size as L3 bc inside the protein is already inside the microsomes. Addition of protease does not change the size of the protein
Lane 5
mRNA + lysate + protease → Nothing on this gel bc w/o microsomes, the protein is left inside the cytoplasm to be digested by protease
Lane 6
mRNA + lysate + microsomes + protease -> Same as L2 bc glycosylation is occurring, sugars are being added so it's heavier than L3 and L4
creds to mbau <3
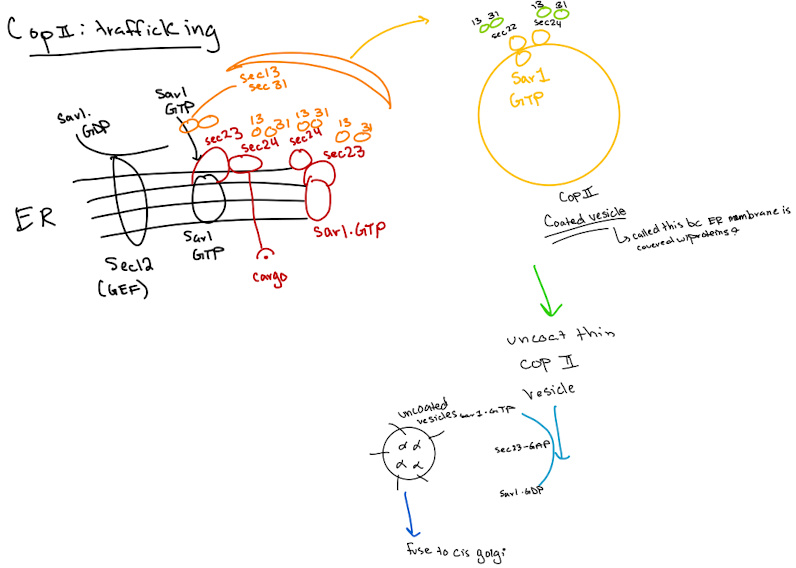
Types of Vesicle Transport
COPII-coated vesicles
COPI-coated vesicles
Cathrin-coated vesicles
COPII-coated vesicles
moves proteins from the rough endoplasmic reticulum to the Golgi apparatus (anterograde transport) “forward”
Cargo Selection
Proteins defined for the Golgi are identified in the ER (ER exit sites)
Coat Protein Assembly (COPII Coat Formation)
Sar1 is activated by GTP and inserts into the ER membrane
Sar1 recruits COPII proteins: Sec 23, Sec24, Sec13, Sec31
Forms coat that bends membrane & helps form vesicle
Vesicle budding
Coated vesicle buds off from ER
Coat Disassembly
After budding, COPII coat is shed, required for docking at Golgi
Vesicle Targeting and Fusion
uncoated vesicle uses SNARE proteins to fuse w/cis-Golgi membrane
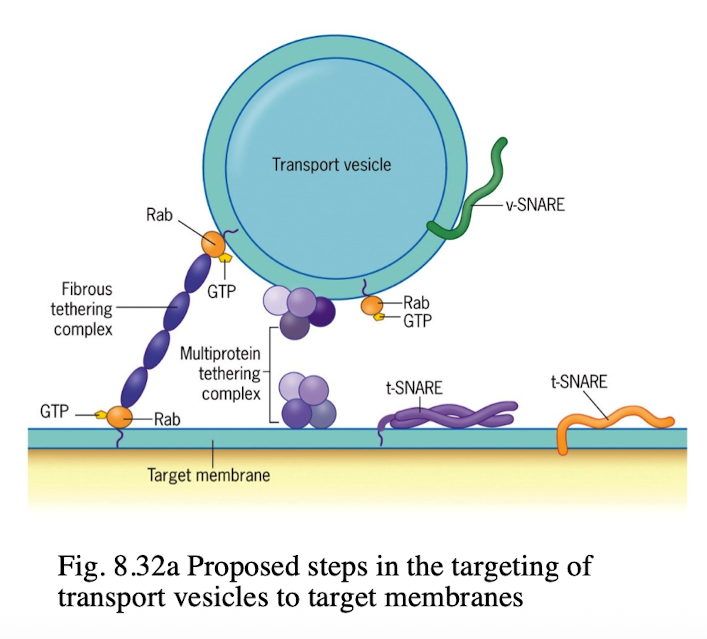
SNAREs (v & t)
integral proteins that bring the vesicle and target compartment in close contact
v-SNAREs are found in transport vesicles, pairs with t-SNAREs to initiate fusion
t-SNAREs are found in target compartments that facilitate vesicle fusion in intracellular transport
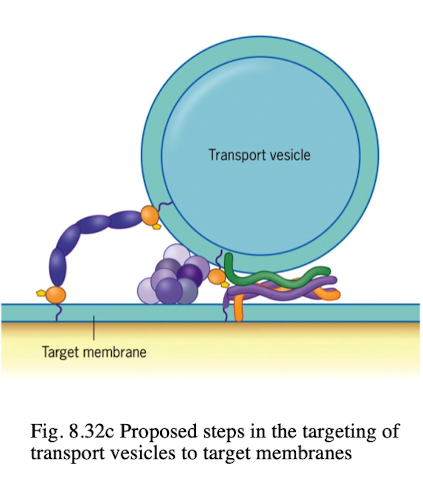
t-SNARE mutant
vesicle docking may occur but fusion fails
Proteins and vesicles accumulate near target membrane
specific effects depend on which t-SNARE is mutated
v-SNARE mutant
vesicle fusion fails
secretory proteins accumulate in vesicles
Blocked secretion or disrupted organelle function
Rabs
proteins that cycle btwn GTP and GDP
GTP-bound Rabs associate w/membranes by a lipid anchor
COPII-uncoated vesicles
Vesicle Formation:
A COP II vesicle forms at the ER membrane, enclosing cargo proteins.
Coat Removal:
The COP II coat disassembles, leaving an uncoated vesicle.
Rab Activation:
Rab-GEF activates Rab-GDP to Rab-GTP on the vesicle.
Tethering:
Rab-GTP on the vesicle interacts with a Rab effector on the cis-Golgi membrane, tethering the vesicle.
SNARE Pairing:
v-SNAREs on the vesicle bind to t-SNAREs on the cis-Golgi membrane, forming a SNARE complex.
Membrane Fusion:
The SNARE complex facilitates fusion of the vesicle membrane with the cis-Golgi membrane.
Cargo Release:
The vesicle's contents are released into the cis-Golgi lumen
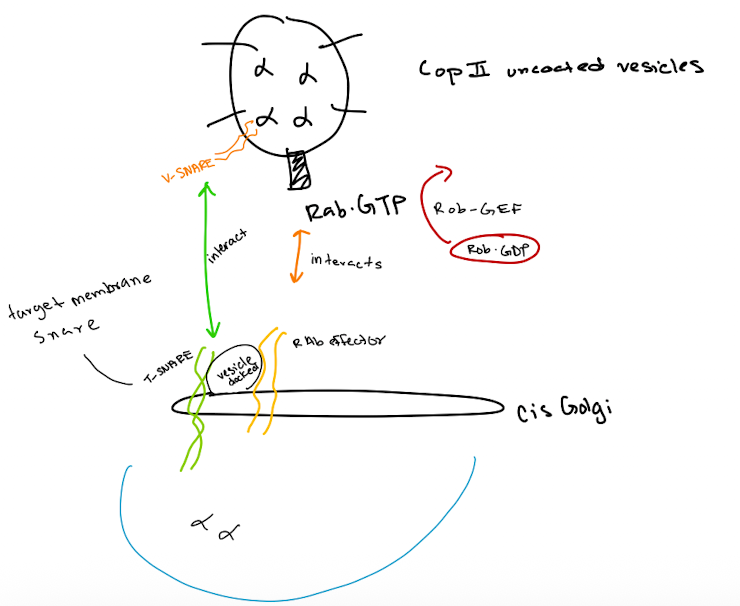
COP1 coated vesicles
For transport from cis-Golgi to ER (retrograde); “backwards”
Maintained by:
Retention of resident molecules excluded from transport vesicles
Retrieval of “escaped” molecules back to normal compartment
Moves Golgi resident enzymes in trans-to-cis direction and ER resident enzymes from ERGIC & Golgi complex back to ER
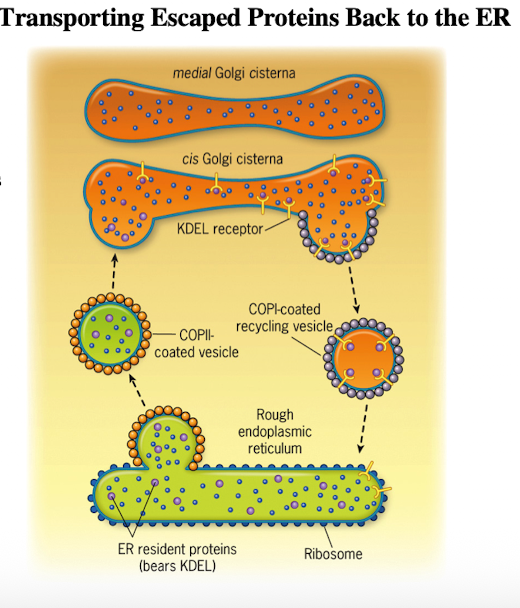
Cathrin-coated vesicles
Essential for selective transport btwn cellular membranes
move materials from the trans-Golgi network (TGN) to endoscopes, lysosomes, and plant vacuoles (plasma membrane)
Endocytosis from plasma membrane
Ran
Is a GTPase that can be bound to GTP or GDP
Ran-GEF act to create a gradient of Ran-GTP in the nucleus
Ran-GAP acts to create a gradient of Ran-GDP in the cytoplasm
Practice Question
Match the statement with the coated protein that it describes.
[Clathrin, COPI, COPII, All of them, None of them]
1. Delivers molecules out of the ER
2. Uses Sar1
3. Uses PI(4,5)P2
4. Used in retrograde transport
5. Make buds that are referred to as pits
6. Can be recruited by coat-recruitment GTPases
7. Interacts with Rabs
8. Recognizes the KDEL sorting signal
9. The outer coat layer is composed of a protein with a triskelion structure of heavy and light chains.
10. The inner coat layer is composed of Sec23/24.
11. Auxilin aids in uncoating.
12. Recognizes cargo receptors with C-terminal KKXX motifs.
1. COPII
2. COPII
3. Clathrin
4. COPI
5. Clathrin
6. All of them
7. None of them
8. None of them
9. Clathrin
10. COPII
11. Clathrin
12. COPI
Practice Question
Select all statements that are TRUE about monomeric GTPases.
1. Rab is lipid-anchored when active.
2. Rab-GAP recruits Rab to the membrane.
3. Arf recruits to Sec13/31.
4. Rab-GTP is bound to GDI.
5. Sar1-GAP leads to vesicle uncoating.
6. Arf is associated with a membrane when it is bound to GTP.
1. Rab is lipid-anchored when active.
6. Arf is associated with a membrane when it is bound to GTP.
Practice Question
Which is false about co-translocation?
1. N-terminal signal sequences are cleaved off via a signal peptidase.
2. Start and stop transfer sequences are indistinguishable.
3. The signal sequence opens the seam of the translocator.
4. BiP helps pull the protein into the lumen of the ER.
5. SRP binds to the signal sequence and the ribosome to pause translation
4. BiP helps pull the protein into the lumen of the ER.
Practice Question
Indicate whether each description better applies to COPI (1), COPII (2), or clathrin (3) coated vesicles. your answer will be a 3 digit number composed of digits 1 to 3 only, eg. 123.
____ Involved in anterograde transport from the ER
____ Involved in transport from the plasma membrane
____ Involved in transport from Golgi to Golgi
2, 3, 1
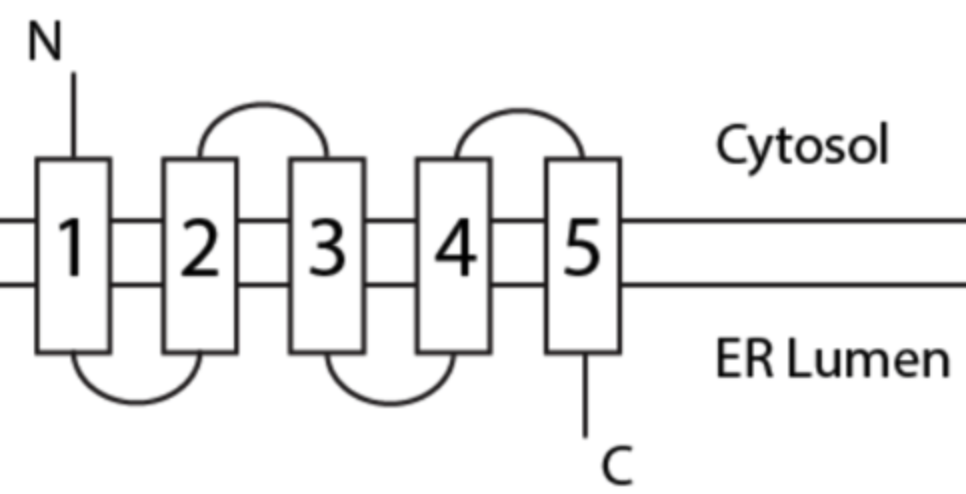
Practice Question
#3 is a start- or stop- transfer sequence?
start