LAB Molecular Diagnosis of Mutation and Inherited Diseases
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
72 Terms
Translocation
Deletion
Insertion
Duplication
Inversion
Tandem Repeat Expansion
Types of Structural Variants:
Point Mutation
Frameshift Mutation
Missense Mutation
Nonsense Mutation
Splice Site Mutation
Types of Mutation:
Destructive (non-balanced)
These SV Types are :
Insertion
Deletion
Interspersed Duplication
Tandem Duplication
Non-Destructive (balanced)
These SV Types are :
Inversion
Translocation (inter- or intra- chromosome)
Structural Variants
Changes occur in our genes, possibly in the sequence of our DNA
The structure of the genetic segment is the one being changed in the chromosome
Keen to excessive mutation
Mutation
Smaller scale changes, possibly only in the DNA sequence.
Inherited disease
Structural Variants and Mutations has similarity because they are both the cause of ________?
Translocation
From one chromosome, there is a segment break as it will attach to another chromosome.
A chromosome arm can be longer compared to the other arm.
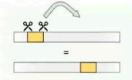
Deletion
Loss of segment
Chromosome or DNA segment shortens.
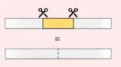
Insertion
Insertion of DNA sequence or segment in a chromosome.
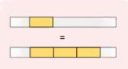
Tandem Duplication
It is adjacent or it came from the original copy.
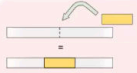
Interspersed Duplication
Possible to be randomly be inserted or duplicated in the chromosome or DNA sequence. It is also not adjacent from the original copy.
Inversion
This is where the segment of DNA happens. Inside the chromosome, the orientation gets flipped which is relative to the normal orientation of the chromosome or DNA sequence.
Tandem Repeat Expansion
There is an expansion in the DNA sequence.
It is known to be Dinucleotide or Nucleotide.
It happens inside the genome.
Complex Rearrangements
Combination of structural variant, possibly 2 or more in 1 variant chromosome.
Point Mutation
Single nucleotide base can be inserted, changed, or deleted from another nucleotide base.
Ex: Sickle Cell Anemia
HPB gene of the beta globulin protein, the beta glutamic acid is being changed.
Sickle Cell Anemia
HPB gene of the beta globulin protein, the beta glutamic acid is being changed
Frameshift Mutation
It can be insertion or deletion, it is being shift into the reading frame of the gene.
Ex: Cystic Fibrosis
If frameshift mutation occurs, transport of chloride ions (page 371)
Cystic Fibrosis
If frameshift mutation occurs, transport of chloride ions
Missense Mutation
DNA changes wherein the codon is being affected. The codon codes for the different amino acids.
Example: Hypercholesterolemia
The LDLR gene is affected as the clearance of LDL (the fats) is being impaired
Hypercholesterolemia
The LDLR gene is affected as the clearance of LDL (the fats) is being impaired
Nonsense Mutation
There are changes or the stop codon is being affected. We will have pre-mature stop codon.
Example: Duchenne Muscular Dystrophy
DMD gene is being affected. (page 371)
Duchenne Muscular Dystrophy
DMD gene is being affected
Splice Site Mutation
The junction of the intron and exon sequence is being affected which also affects the mRNA splicing.
Example: Beta-Thalassemia
HPB gene is affected as there would be abnormal splicing. Decrease or reduced functional hemoglobin, (page 376)
Beta-Thalassemia
HPB gene is affected as there would be abnormal splicing. Decrease or reduced functional hemoglobin.
Diseases Caused by Genetic Mutation and Structural Variant
THESE ARE:
Alpha-1 Antitrypsin Deficiency
Cystic Fibrosis
Duchenne Muscular Dystrophy
Huntington’s Disease
Sickle Cell Disease
Thalassemia
Tay-Sachs Disease
Alcohol Dependence
Charcot-Marie-Tooth Disease
Cardiovascular Disease (CVD)
Parkinson’s Disease
Achondroplasia
Duane’s Retraction Syndrome
Fragile X Syndrome
Hemophilia
Marfan Syndrome
Cri du chat Syndrome
Dominant Optic Atrophy (DOA)
Achondroplasia
What Disease has this type of Gene Involved?
GENE: FGFR3 gene
Duane’s Retraction Syndrome
What Disease has this type of Gene Involved?
GENE: CHN1 gene
Fragile X Syndrome
What Disease has this type of Gene Involved?
GENE: FMR1 gene
Hemophilia A
What Disease has this type of Gene Involved?
GENE: F8 gene
Hemophilia B
What Disease has this type of Gene Involved?
GENE: F9 gene
Marfan Syndrome
What Disease has this type of Gene Involved?
GENE: FBN1 gene
Cri du chat Syndrome
What Disease has this type of Gene Involved?
GENE: Deletion of part of the short (p) arm of chromosome 5
Dominant Optic Atrophy (DOA)
What Disease has this type of Gene Involved?
GENE: OPA1 gene
Alpha-1 Antitrypsin Deficiency
What Disease has this type of Gene Involved?
GENE: SERPINA1 gene
Cystic Fibrosis
What Disease has this type of Gene Involved?
GENE: CFTR gene
Duchenne Muscular Dystrophy
What Disease has this type of Gene Involved?
GENE: DMD gene
Huntington’s Disease
What Disease has this type of Gene Involved?
GENE: HTT gene
Sickle Cell Disease
What Disease has this type of Gene Involved?
GENE: HBB gene
Thalassemia
What Disease has this type of Gene Involved?
GENE: HBB gene, HBA1 and HBA2 genes
Tay-Sachs Disease
What Disease has this type of Gene Involved?
GENE: HEXA gene
Alcohol Dependence
What Disease has this type of Gene Involved?
GENE: Multiple gene involved, including ADH1B and ALDH2 genes
Charcot-Marie-Tooth Disease
What Disease has this type of Gene Involved?
GENE: Multiple genes, including PMP22, MFN2, and others
Cardiovascular Disease (CVD)
What Disease has this type of Gene Involved?
GENE: APOE, LDLR
Parkinson’s Disease
What Disease has this type of Gene Involved?
GENE: PARK2, SNCA, LRRK2, and other genes
1. The current knowledge of the gene(s) associated with the disease in question.
2. Degree of Molecular Heterogeneity
There are 2 Factors to consider in Choice of testing, these are:
The current knowledge of the gene(s) associated with the disease in question.
These are part of what factor in choice of testing?
Those for which the causative gene has been isolated and
Those for which it has not.
We can know it by checking the family history.
If the person has been tested by the signs and symptoms.
We can have an initial diagnosis or presumption regarding the disease the patient faces.
Once the disease is isolated, we can use direct mutation analysis .
PCR
NGS
If hindi pa naiisolate or naiidentify what disease, we use unknown or non-isolated pack .
We use linkage analysis as we use a marker known as STR and SNP.
Short Tandem Repeats (STR)
Single Nucleotide Polymorphism (SNP)
PCR
NGS
Once the disease is isolated, we can use Direct Mutation Analysis. And Examples are:
Short Tandem Repeats (STR)
Single Nucleotide Polymorphism (SNP)
If hindi pa naiisolate or naiidentify what disease, we use Unknown or Non-Isolated pack. And Examples are:
Degree of Molecular Heterogeneity
These are part of what factor in choice of testing?
If there are multiple genes involved that cause the disease, usually pag mas maraming genes ang involved - mas maraming test ang gagawin.
Direct Mutation Analysis
most preferred than the direct linkage analysis.
Not timing consuming since you already know the disease.
You can directly diagnose it.
Direct Mutation Analysis
must preferred than the direct linkage analysis.
Not timing consuming since you already know the disease.
You can directly diagnose it.
PCR Amplification
Gel Electrophoresis
Restriction Enzyme Analysis
Dot Blot Hybridization
Microarray Analysis
Examples of Direct Mutation Analysis Application:
PCR Amplification
To amplify our products.
To produce multiple copies.
Gel Electrophoresis
For reading the different fragment size of the DNA.
We compare it to the control (normal or diseased)
Interpretation: If there are changes in the band. There is a mutation in the patient.
Insertion - additional band compared dun sa control.
Deletion - missing band.
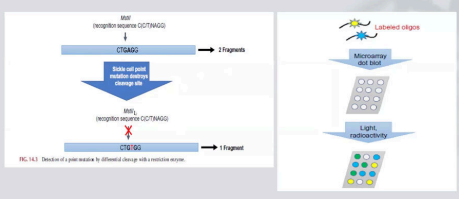
Restriction Enzyme Analysis
We would see if the restriction enzyme would react - if there is a possibility that it contains patient DNA with restriction site.
If the restriction enzyme would react with the restriction site through cleavage pattern.
Dot Blot Hybridization
We use labelled oligonucleotides which are fluorescently labelled.
Microarray Analysis
We can detect multiple mutations simultaneously.
We use multiple probes to cover the whole genome as we can consider the ACGH or ECGH
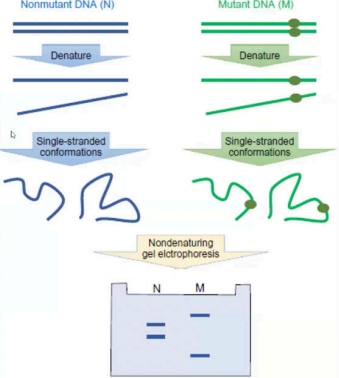
Single-Strand Conformation Polymorphism (SSCP)
Denaturing Gradient Gel Electrophoresis (DGGE)
Denaturing High-Performance Liquid Chromatography (DHPLC)
Examples of Techniques used in Unknown Mutation:
Chemical Base
Heat Base
Temperature Base
2 Methods for DDG:
DHPLC
we use chromatography column
We look for the difference in retention time (gaano katagal madedenature)
We use PCR amplification.
Wild Type DNA - standard and reference.
Mutant
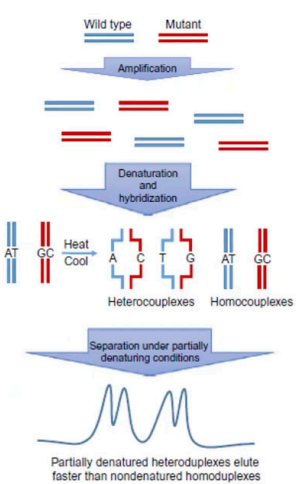
Heterocouplexes
nagband yung wild type and mutant. Nahybridize sila pareho.
Homocouplexes
it came from the original double stranded DNA.
Multiplex Ligation-Dependent Probe Amplification (MLPA)
Capillary Electrophoresis-Strand Conformation Polymorphism (CE-SCP)
Stuffer-free Multiplex CNV Detection Method
Examples of Techniques used in Detecting Structural Changes:
Multiplex Ligation-Dependent Probe Amplification (MLPA)
It is a method used to detect copy number variations (CNVs) in the genome.
It combines the advantages of multiplex PCR and capillary electrophoresis and can detect deletions, duplications, or insertions in specific genomic regions.
Duchenne Muscular Dystrophy
Prader-Willi Syndrome
Angelman Syndrome
Diseases tested using MLPA include the following:
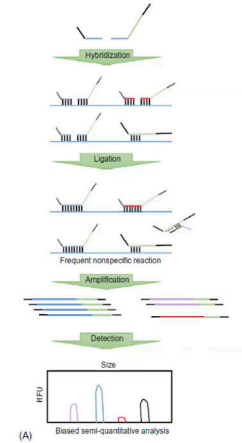
Conventional MLPA
The picture is the flow of what MLPA?
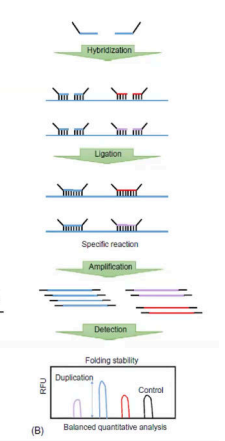
Stuffer-free MLPAS
The picture is the flow of what MLPA?
Array Comparative Genomic Hybridization (aCGH)
Next-Generation Sequencing (NGS)
Examples of Whole Genome Analysis Techniques:
Array Comparative Genomic Hybridization (aCGH)
We use so many specific probes as it will cover the whole genome to detect.
The probes will bind to the DNA.
Data Analysis: if there would be fluorescence in the genome.
Next-Generation Sequencing (NGS)
More rapid and cost-effective.
Detects structural variation.
Higher solution.
We could analyse specific segment or site of the genome that we want to test.
Single-cell sequencing (SCS)
is a cutting-edge molecular technique that allows for the analysis of individual cells at the genomic, transcriptomic, or epigenomic level.
Unlike traditional bulk sequencing methods, which analyze populations of cells together, SCS provides insights into the genetic molecular characteristics of individual cells within a heterogenous sample.
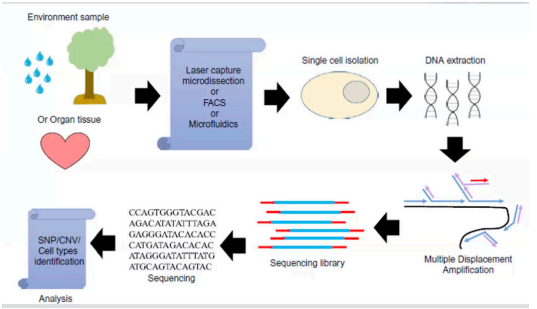
Cancer Research
Embryonic Development
Clinical Diagnostics
Stem Cell Research
Epigenetics
Single Cell Sequencing is used in the ff: