cell biology exam 1
1/124
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
125 Terms
fundamental properties that characterise living things and distinguish them from nonliving matter
organization, metabolism, homeostasis, growth, reproduction, response, adaptaition
cell biology
the study of cells and their structure, function, and behavior
what determines the nature of the organism developing from a cell
hereditary information in the fertilized egg cell
how many described species living on earth today
> 2 million
all cells store their heredetary information in the form of
double stranded DNA molecules
all cells replicate their hereditary information by
templated polymerization
building block of DNA
nucleotides
DNA is made up of
sugar-phosphate + base
what makes the DNA strand backbone directional/polar & why does this matter
asymmetric sugar-phosphate units
guides processes by which the information in DNA is interpertedd and copied. read left to right
templated polymerization
polymer is synthesized by using an existing template to ensure complementary base pairing
ex- DNA replication, transcription (DNA → RNA), and translation (RNA → protein)
transcription
DNA → RNA
translation
RNA → protein
what molecular principle underlys templated polymerization
complementary base pairing (nucleic acids) or codon-anticodon pairing (translation)
hydrogen bonds
hold complementary bases together (A-T) (C-G)
bond between two strands
covalent bonds
connect sugar, phosphate, and nitrogenous base
bond within the strand
DNA double helix
two DNA strands twist around each other to form this structure that can accomodate any sequence of nucleotides without altering its basic double-helical structure
DNA replication
double helix unwinds and each parent strand is a template for a new strand.
DNA replication (detailed)
Helicase unwinds DNA, primase lays down RNA primers, DNA polymerase synthesizes new DNA 5′ → 3′, the leading strand is continuous while the lagging strand forms Okazaki fragments, and DNA ligase seals the gaps. Proofreading ensures high fidelity.
process of DNA→ amino acids
DNA replication
Transcription- gene is copied into mRNA
Translation- RNA molecule guides protein synthesis where amino acids are made
sugar is a subunit for which macromolecule
polysaccharide
amino acid is a subunit for which macromolecule
protein
nucleotide is a subunit for which macromolecule
nucleic acid
% volume of a cell
70% H2O, 30% chemicals
how is life an autocatalytic process
DNA and RNA provide nucleotide sequence info used to produce proteins and copy themselves
Proteins provide catalytic activity needed to synthesize DNA, RNA, and proteins
These feedback loops create self replicating system allowing them to reproduce
phospholipid molecule
major component of cell membranes. contains a hydrophilic phosphate group head and two hydrophobic fatty acid tails
behavior of phospholipid molecules in water
aggregate to form lipid bilayers that fold in on themselves to form sealed compartments known as vesicles
tails look like they are touching each other
behavior of phospholipid molecules between water and oil
arrange themselves as a monolayer with head group facing water and tail facing oil
cells can exist with _ genes
>500
all cells…
store hereditary information in form of dsDNA molecules
replicate hereditary information through templated polymerization
transcribe portions of their DNA into RNA molecules
translate RNA into protein
enclosed in a plama membrane which nutrients and waste must pass
three major domains in the tree of life
eukaryotes, bacteria, archaea
most diverse group of organisms on the planet
bacteria
gram positive
bacteria that lack an outer membrane
gram negative
bacteria that have an outer and inner (plasma) membrane
most mysterious domain of life
archaea
only shwon to be a seperate domain through DNA analysis in 1977
most thrive under extreme conditions
total biomass on earth
~550 Gt C, with plants being about 450 of that
intragenic mutation
mutation that occurs within a single gene

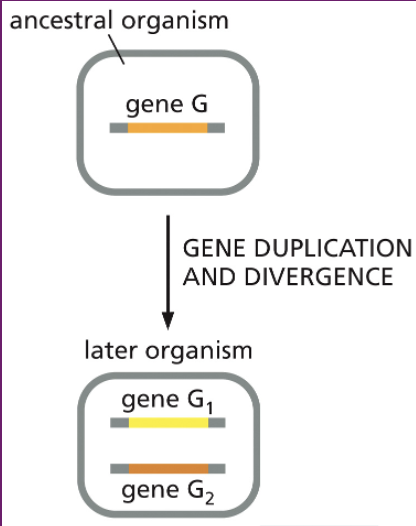
gene duplication
segment of DNA containing a gene is copied one or more times in the genome.

DNA segment shuffling
segments of DNA within or between genes are rearranged, creating new combinations of exons

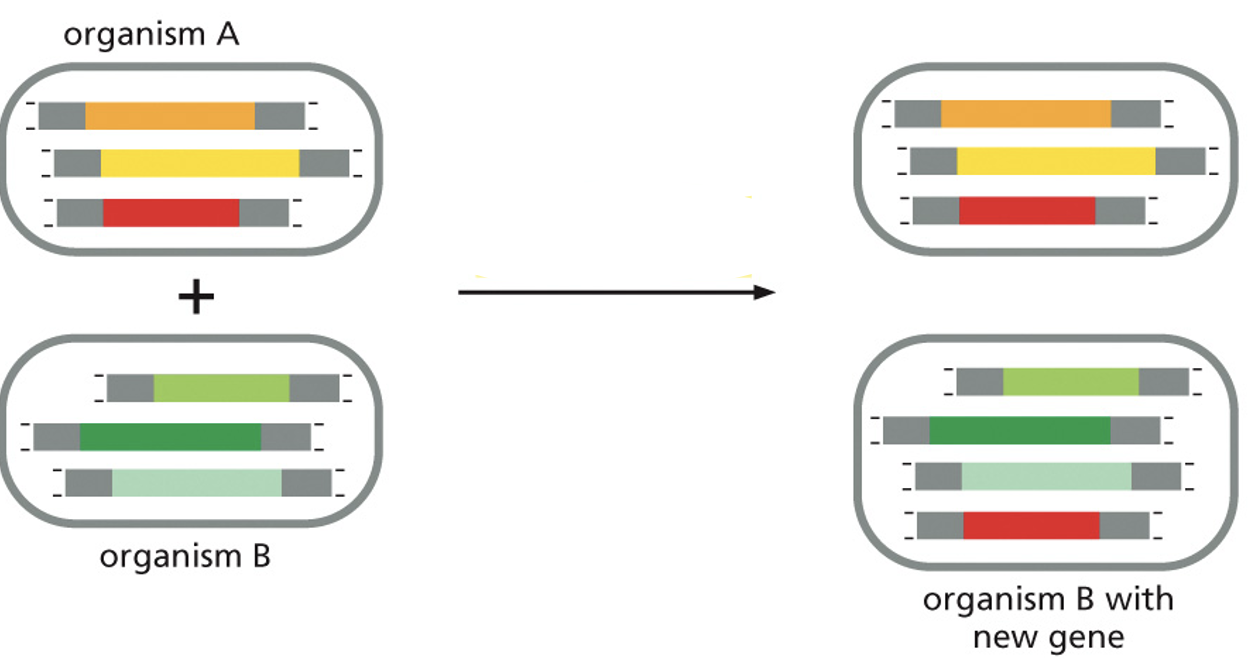
horizontal DNA transfer
movement of genetic material between organisms that are not parent and offspring, commonly in bacteria via transformation, transduction, or conjugation
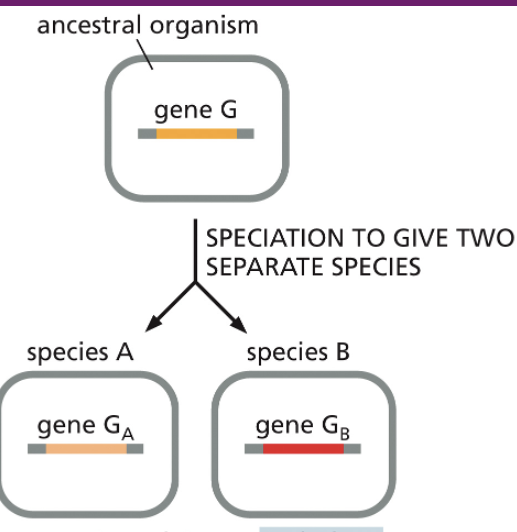
orthologs
genes in different species that evolved from a common ancestral gene; usually retain the same function.
Human β-globin gene vs. mouse β-globin gene (both carry oxygen in red blood cells).
paralogs
genes within the same genome that arose from gene duplication; can evolve new functions.
ex- Human α-globin vs. β-globin genes (both part of hemoglobin, but slightly different roles).
what are genes made up of in eukaryotes
promotor regions and alterning introns (noncoding) and exons (codig) regions
steps to produce a protein
transcription of gene from DNA to RNA
removal of introns and splicing of exons
translation of spliced RNA into chain of amino acids
posttranslational modification of the protein molecule e
endocytosis
the process in which eukaryotic cells import extracellular materials
exocytosis
the process by which eukaryhotic cells secrete intracellular materials
membrane enclosed organelles in the eukaryotic cell
lysosome, mitochondria, peroxisome, golgi apparatus, endoplasmic reticulum, endosome, transport vesicle, nuclear envelope, lysosome
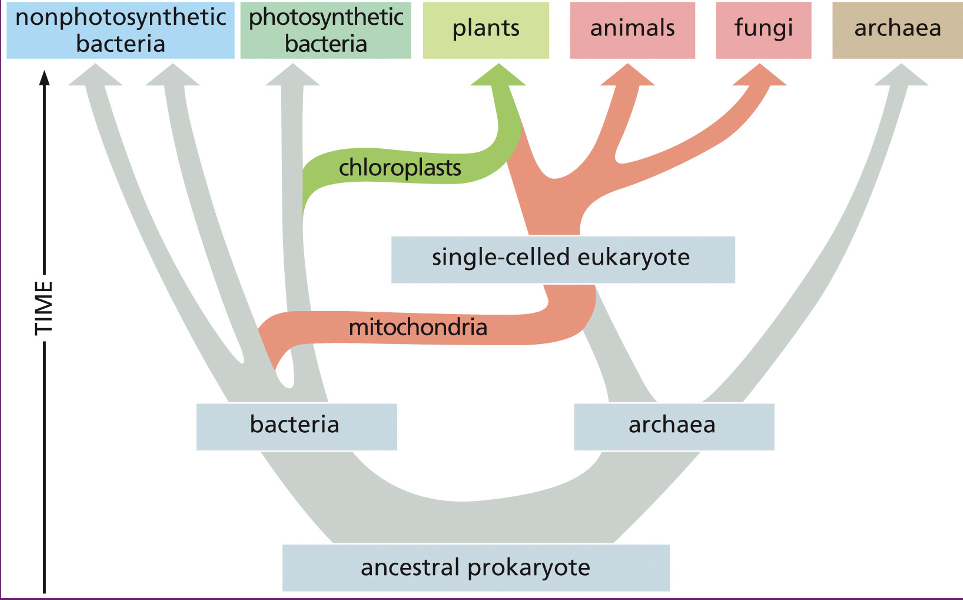
how did mitochondria evolve
symbiotic bacterium captured by an ancienet archaeon
possible steps in eukaryotic cell evolution
enclosure of ectosymbiont by archaeal mebrane fusion and then the escape of the endosymbiot into cytosol and formation of new intracellular compartments
where did chloroplasts evolve from
symbiotic photosynthetic bacterium engulfed by an ancient eukaryotic cell
evolution of cells in the tree of life
hybrid genomes
genome that contains genetic material from two distinct sources, often resulting from hybridization between species or strains
all living cells are thought to have evolved from
an ancestral prokaryotic cell (3.5 BYA)
do different cell types in a multicellular organism contain different DNA?
No, all cell types contain the same DNA, even though they perform different functions.
How do different cell types produce different proteins and RNAs?
Different cell types synthesize different sets of RNAs and proteins, even though they have the same DNA.
How can the spectrum of mRNAs in a cell be used?
(which genes are transcribed) can be used to accurately identify the cell type.
How can external signals affect a cell?
External signals can cause a cell to change the expression of its genes, adapting to new conditions.
at what steps can gene expression be regulated?
gene expression can be regulated at many steps along the pathway from DNA → RNA → Protein.
What does RNA-seq measure in cells?
RNA-seq measures the amount and sequence of RNA in cells by converting RNA into fragments, sequencing them, and mapping the reads to the genome
In RNA-seq data, what does the height of the trace represent?
the number of RNA sequence reads that match a particular region of the genome.
what key concept does RNA-seq illustrate about multicellular organisms?
All cells have the same DNA, but gene expression varies by gene and tissue, explaining differences in cell type and function.
places at which eukaryotic gene expression can be controlled
transcription
RNA processing
RNA transport and localization
translation
mRNA degradation
protein degradatin
protein activity
why does dimerization of transcription regulators matter?
it increases both the affinity (strength of binding) and specificity (recognition of the correct DNA sequence) of transcription regulators for DNA.
cis-regulatory sequence
DNA region near a gene where transcription regulators (transcription factors) bind to control gene expression. typically embedded within longer DNA molecules and regulate nearby genes.
dimer
a pair of transcription regulators (either identical or different) that join together to bind DNA with increased affinity and specificity.
homodimer
complex formed by two identical transcription regulator proteins that bind DNA together
heterodimer
complex formed by two different transcription regulator proteins that bind DNA together.
transcription regulator
protein that binds specific DNA sequences (cis-regulatory elements) to control gene expression.
how does the bacterial transcription regulator NtrC activate transcription?
NtrC binds a distant DNA site and activates transcription by contacting RNA polymerase, with the intervening DNA looped out to bring them together.
DNA looping
occurs when a regulatory protein binds a distant site and the DNA loops out so the protein can interact with RNA polymerase or other transcription machinery, influencing gene expression.
how do transcription activators work synergistically
multiple transcription activators can work together, producing a combined effect on gene expression that is greater than the sum of their individual effects.
primary cells
directly isolated from tissue; represent in vivo physiology; limited growth potential.
immortal cell line
a population of cells that can divide indefinitely in culture, bypassing the normal limit on cell divisions
immortalized (by mutation or deliberate modification); robust and easy to grow; unlimited growth; may have altered physiology and can change over time with passaging.
SDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis)
technique in which proteins are denautred and coated with negativley charged SDS then separated in a polyacrylamide gel based on size. used to estimate protein molecular weight and analyze subunit composition
Western Blotting
detects specific proteins in a sample using antibodies. proteins are separated by SDS-PAGE, transferred to a membrane, then probed with antibodies to identify protein presence, size, and abundance
immunofluorescence (IF)
technique that uses fluorescently labled antibodies to detect and visualize specific proteins or antigens in cells or tissues to determine their location, abundance, and distribution
x-ray crystallography
technique that determiens the three-dimensional atomic structure of a molecule by anaylizing th ediffraction pattern produced when x-rays pass through a crystal of the molecule. commonly used for proteins, nucleic acids, and small molecules
how do restriction nucleases cut DNA, what is their importance to
recognize DNA sequences, act as dimers, and cut both strands either blunt-ended or staggered, sticky ends. sticky ends allow fragments to rejoin via complementary base-pairing.
sticky ends
result from the cleaveage of DNA by restriction nucleases and allow DNA fragments to rejoin via complementary base pairing
main steps for isolating DNA from cells
cell lysis- break open cells while keeping nuclei intact
nucleus separation- isolate nuclei via centrifugation or filtration
nuclear lysis- break open nuclei to release DNA
purification- remove proteins and RNA
DNA Library
collecition of DNA fragments stored in host cells where each fragment is cloned into a vector, representing the genome
DNA library
A collection of DNA fragments representing an entire genome, stored in host cells or in vitro. made by fragmenting DNA, cloning fragments into vectors and introducing them into host cells, then this collection of host cells represents the whole genome.
hybridization
he process in which two single-stranded DNA molecules with complementary sequences pair up to form a double-stranded structure. This process can occur after DNA has been separated by heat or chemical treatment, allowing the strands to find and bind their matching sequence.
FISH (Fluorescence In Situ Hybridization)
uses labeled DNA probes that hybridize to their complementary sequences on partially denatured chromosomes. The probes are detected with fluorescent antibodies, allowing visualization of specific DNA regions. In metaphase chromosomes, each replicated chromosome contains two identical DNA helices, so each probe produces two fluorescent signals per chromosome.
PCR (polymerase chain reaction)
technique used to amplify specific DNA sequences in vitro. It uses DNA primers, a thermostable DNA polymerase, and repeated cycles of heating and cooling to denature DNA, allow primers to bind, and synthesize new DNA, producing millions of copies of the target sequence.
what are the steps/cycles of PCR
denaturation, where double-stranded DNA is heated to separate the strands; annealing, where primers bind to their complementary sequences on the single-stranded DNA; and extension, where DNA polymerase synthesizes new DNA. These steps are repeated 25–35 times, doubling the DNA each cycle, producing millions of copies of the target sequence.
How is PCR used in forensic science with STRs to identify individuals?
PCR is used to amplify short tandem repeats, repetitive DNA sequences that vary between individuals.
primers flank each STR locus, producing DNA fragments which are separated by gel electrophoresis and produce a band pattern, creating a dna fingerprint. this technique is used in paternity testing
how is PCR used for DNA fingerprinting
Targets variable regions of DNA called VNTRs or STRs, which differ between individuals. Amplifying and analyzing these regions produces a distinctive DNA pattern for each person, allowing individuals to be distinguished (except identical twins).
wild type
normal version of a gene that produces the typical functional protein found in most individuals
loss-of-function mutation
reduces or limits protein activity. can occur via point mutation (single base change), truncation (premature stop codon), or deletion (removal of part or all of the gene)
conditional loss-of-function mutation
produces a protein that is functional only under certain conditions such as specific temperatures or environmental factors
gain-of-function mutation
enhances the proteins ability or gives it a new function, possibly leading to dominant phenotypes or abnormal activity
how can engineered genes be turned on and off with tetracycline (Tet) systems
genes are controlled using the repressor fused to a transcription activator. In the absence of doxycycline, the gene is expressed; in its presence, the repressor dissociates, turning the gene off. Tissue-specific control is possible with a tissue-specific promoter for the repressor.
how is CRISPR-Cas9 used to study and edit genes
uses the Cas9 protein bound to a guide RNA to target specific genomic sequences adjacent to a PAM motif. Cas9 can create double-strand breaks, which are repaired by nonhomologous end joining (disrupting gene function) or homologous recombination with an altered template (precise edits). Mutant Cas9 that cannot cut DNA can activate or repress genes.
Originally an adaptive immune system in bacteria, CRISPR-Cas9 was adapted in 2012 for genome editing and recognized as a major breakthrough in gene modification.
RNA interference (RNAi)
a type of Post-Transcriptional Gene Silencing (PTGS), natural mechanism where dsRNA triggers sequence-specific gene silencing. helps cells resist parasitic or pathogenic nucleic acids and regulate protein-coding genes.
in situ hybridization
a technique that uses labled nucleic acid probes to detect specific mRNA sequences within inact cells or tissues, revealing spatial patterns of gene expression
ex- in Drosophila, probes reveal eve (magenta) and ftz (green) stripes in early embryoes and DNA is counterstained (whtie) to mark nuclei
Whole-mount immuno-coupled hybridization chain reaction (WICHCR)
enables high-resolution detection of mRNA and protein in inact tissues
protein sequencing
identifying the precise order of amino acids in a protein or peptide, essential for characterizing proteins, understanding function, and identifying post-translational modifications. two primary methods are mass spectrometry and edman degradatin
protein sequencing by mass spectrometry
measures mass-to-charge ratio of ionized peptides and can be used to identify, quantify, and characterize proteins, including post-translational modifications. three main approaches- bottom-up, middle-down, and top-down
bottom-up proteomics
digestion, small peptides to be analyzed
proteins are digested into short peptides (usually with trypsyn) before mass spectrometry analysis. most common method, good for large-scale protein id and quantification, but may loose some info on full protein sequence or post-translational modifications
middle-down proteomics
digestion, large peptides to be separated and analyzed
proteins are cleaved into larger fragments (not full digestion) before mass spectrometry. balances coverage of post-translational modifications and sequence information with manageble fragment size.