Lecture 2: Bacterial transcription and regulation
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
88 Terms
Transcription
process in which RNA is synthesized on the DNA template by RNA polymerase
RNA polymerase similarities to DNA polymerase
synthesis of new chain guided by template, 5’ to 3’ synthesis, and adds rNTPs instead of dNTPs to the chain
RNA polymerase differences to DNA polymerase
no need for primer (de novo synthesis), not restricted to S-phase, only uses template DNA strand for transcription, more error prone (1000x less accurate), different genes are transcribed at different times with different efficiency
Operon
transcription unit of prokaryotes. Within a single operon there are several structural genes. Has a promoter before (upstream) the genes and a terminator after (downstream) the genes.
Upstream vs downstream
"upstream" is towards the 5' end and "downstream" is towards the 3' end
Everything before the operon start is denoted as —- of the operon
upstream
The 1st nucleotide of an operon is denoted as what?
+1
Everything upstream of an operon is labeled as what?
a negative number (ex -1 is the first nucleotide before the gene)
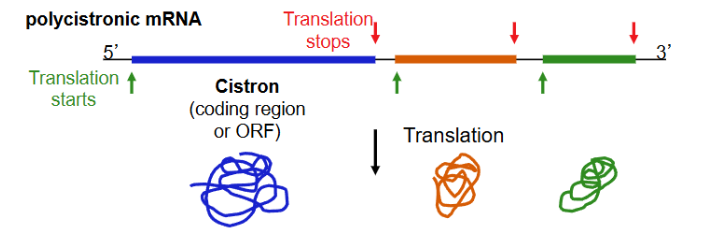
Polycistronic mRNA
singular RNA with multiple protein coding regions (example here has code from three different genes each with their own translation start sites)
How many RNA polymerases do bacteria have?
a single polymerase
Subunits that make the core RNA polymerase enzyme in bacteria
2 alpha(𝛼), 1 beta(β), 1 beta prime(β’), and 1 omega (ω)
Bacterial core RNA polymerase may bind to DNA without a sigma subunit in what conditions?
nicked DNA or the end of a double stranded DNA
Sigma(σ) subunit
not part of the bacterial core RNA polymerase. Responsible for recognizing the promoter.
Different — recognize different promoters in bacteria
sigma subunits
RNA polymerase in bacteria is made up of what two things?
sigma subunit and the core enzyme
t/f the number of sigma factors a bacteria has is the same between all species
false
Bacteria have multiple sigma factors in order to
recognize multiple promoters

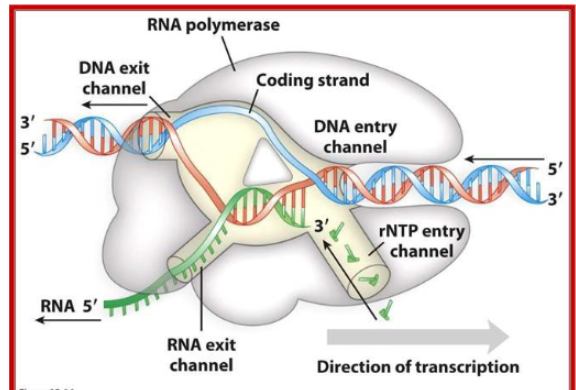
Label this bacterial RNA polymerase
(use 2d image from slides)
Initiation (overview of steps and complex name at each step)
RNA polymerase recognizes and binds the promoter via the sigma factor (closed complex). RNA polymerase unwinds DNA to form the transcription bubble (open complex). The first phosphodiester bonds between rNTPs are made (unstable ternary complex). The sigma factor is released after about 10 nucleotides are added (stable ternary complex)
Closed complex (initiation)
sigma factor of the RNA polymerase recognizes the conserved sequences at -35 and -10 region and binds the RNA polymerase holoenzyme to the DNA. DNA is not yet unwound.
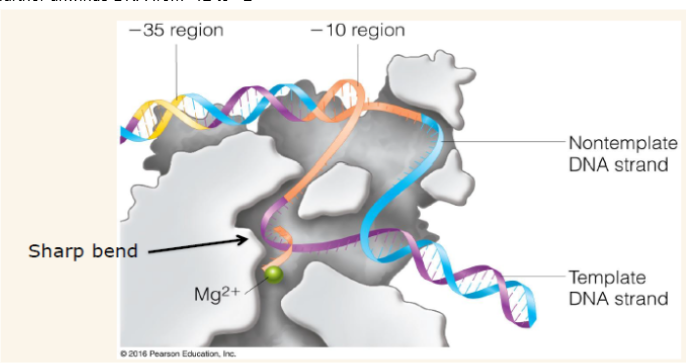
Open complex (initiation)
when DNA strands are separated after the RNA polymerase has bound to the DNA. -10 region is unwound then a Mg2+ dependent isomerization further unwinds DNA from -12 to +2
Unstable ternary complex (initiation)
when the first phosphodiester bonds between rNTPs are made, forming an unstable ternary (DNA, RNA pol, and RNA) complex. Abortive initiation occurs here due to sigma factor pulling RNA pol back to promoter and partially blocking the RNA exit channel.
Stable ternary complex (initiation)
after about 10 nucleotides are added to the nascent RNA strand, the sigma factor is released from the RNA pol enzyme and a stable ternary complex forms after promoter escape
Pribnow box
aka -10 region. AT rich region where DNA is first unwound for transcription. One of two consensus sequences recognized by the sigma factor
What can make a promoter “weaker”
differences from the consensus sequences at the -35 and -10 regions that its sigma factor recognizes
In e.coli what two conserved sequences are found at the -10 region and -35 region respectively for a promoter recognized by sigma 70?
-10-TATAAT; -35- TTGACA
N17 region
region found between the -35 region and -10 region. Sequence does not matter but number of nucleotides in this region can affect sigma factor binding
Sigma 32 binds to genes involved with bacterial response to what?
heat shock
UP element
upstream promoter element. AT rich region Recognized by RNA polymerase itself (via C terminal domain of alpha subunit). Strengthens the promoter and is present is in highly expressed in genes
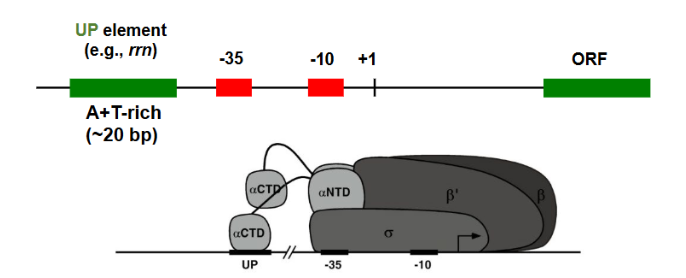
Sigma factors binds to DNA through the —-
major groove
DNA footprinting analysis
Used to identify any protein binding sites of DNA and RNA. label DNA on one strand and then let the protein of interest bind to the DNA (also use another unbound DNA as a control). Cut each DNA strand with a nonspecific DNAse (will cut DNA in spots it can reach. It won't cut where the protein is bound).
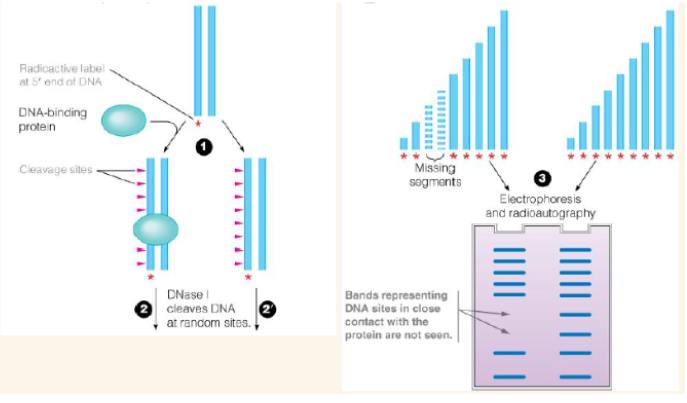
2 methods used in order determine conserved sequences bound by sigma factors
DNA footprinting and/or chip-seq
Abortive initiation
the first rounds of transcriptions are non-productive. Short strands of RNA are synthesized and released repeatedly before productive transcription starts. Caused by sigma factor partially blocking RNA exit channels and pulling the RNA polymerase back to the consensus sequences. Short strands of RNA (~10 nt) synthesized and released until promoter escape
Promoter escape
end of abortive initiation. Formation of stable ternary complex.
Elongations overview
chain elongation continues in 5’ in 3’ directions around 40-80 nt/s.
—-- forms ahead and —-- forms behind the transcription bubble which is resolved by —-
positive supercoils; negative supercoils; topoisomerases
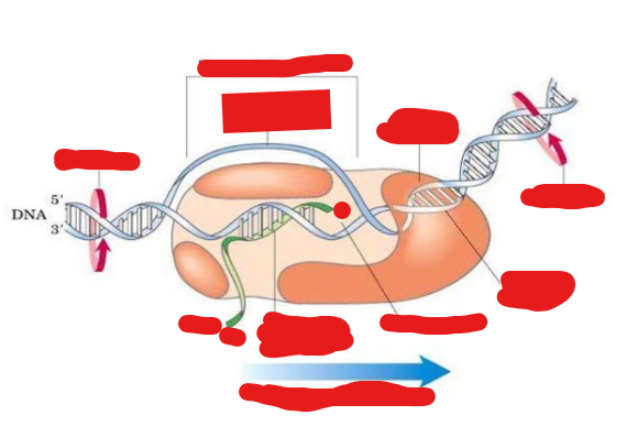
Labels this transcription elongation complex
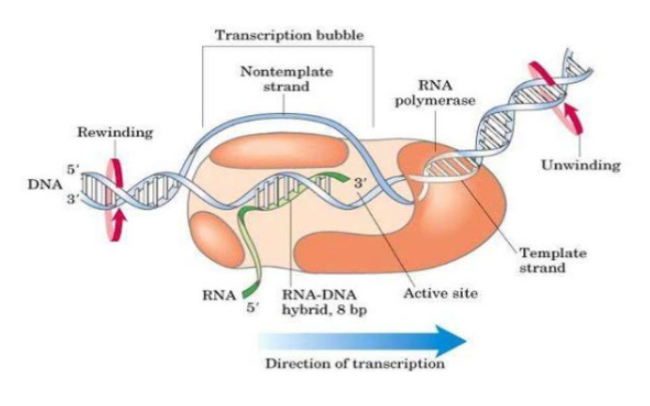
Average elongation rate of bacteria
45 nt/s
Maximal elongation rate of bacteria
80 nt/s
What does transcription elongation being processive means?
RNA polymerase does not come off the DNA until termination
There is about —- nt within RNAP at as the ternary complex
14
Reasons RNA polymerase may need to pause
RNA secondary structures, difficult sequences(ie GC rich regions), backtracking, and limiting NTP
t/f RNAP does not have proofreading activity, which is why it is much more error prone than DNA pol
false, while it is more error-prone, it still has proofreading activity
How may RNAP proofread
either through an endonuclease that travels with it or by reversing the reaction between the last nucleotide that was added to the RNA and the RNA molecule
Transcription in bacteria is coupled with —--
translation
When does translation begin on a mRNA strand?
as it is being transcribed, translation begins occurring on the nascent strand
Transcription termination
transcription stops after terminator sequence. RNA transcript is released and RNA polymerase detaches. May be rho-dependent or intrinsic (rho-independent) termination
Intrinsic termination
DNA template contains GC rich inverted repeats followed by about 8 A which creates a hairpin loop in the RNA strand. This stalls the RNA polymerase and destabilizes the 5’ end of the hybrid RNA RNAP complex. The RNA/DNA duplex (AU bonds) is weakened so RNA transcript is released.
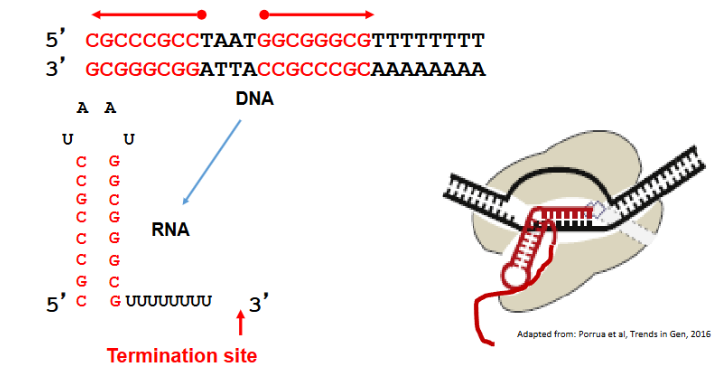
Most termination in bacteria is via —- termination
intrinsic
Rho-dependent termination
Rho factor binds to a specific sequence on nascent RNA (rut site). Rho factor moves 5’-3’ on transcript hydrolyzing ATP. stalling of polymerase facilitates rho termination. When polymerase stalls due to hairpin or GC rich sequence, the rho-factor can catch up and melt the RNA/DNA hybrid, releasing the transcript.
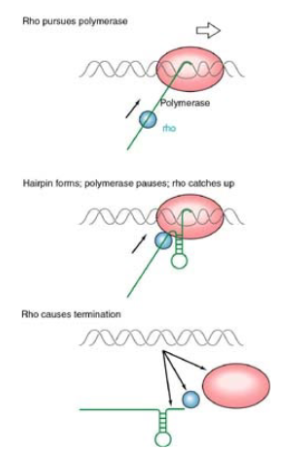
Rho-factor
ATP-dependent helicase that unwinds DNA/RNA hybrids and causes termination. Moves in 5’ to 3’ direction
Rho utilization site (rut site)
~40 nt C-rich sequence to which rho-factor binds and begins moving along the RNA transcript in order to cause termination
Why is post-transcriptional modification in bacteria relatively rare?
due to coupling of translation with transcription
All rRNA are derived from —- in prokaryotes
a single precursor which is cleaved into the separate rRNA molecules
polyA facilitates what in bacteria?
RNA degradation
t/f post-transcriptional processing of rRNA in bacteria is extensive
true
Bacterial mRNAs are (stable or unstable) with half-lifes measured in —--
unstable; minutes
RNA degradosome
complex of RNAses and RNA helicases which helps in degradation of different RNAs
RNAs in bacteria are degraded by —- and —-
endonucleases and exonucleases
Why does adding a poly-A tail on the 3' end of bacterial RNAs make them more unstable?
3’ exonucleases require a single stranded RNA to begin chewing up RNA. Most RNAs end with a hairpin loop due to transcription termination which blocks the exonucleases from beginning in degradation. The poly A tail gives the exonucleases a place to begin degradation.
Constitutive expression
expressed all the time
Repressible gene
normally on, expression may be turned off by repressor
Inducible gene
normally off, expression can be turned on (induced) by an inducer)
On what three levels may gene expression be regulated in bacteria?
transcription, mRNA stability, and translation
What is the most important regulation mechanism for gene expression in bacteria
regulation of transcription
What type of system and regulation does the lac operon fall under?
inducible and positive and negative regulation
Show how lactose is metabolized in bacteria

lacZ
part of lac operon. encodes beta-galactosidase which cleaves lactose into galactose and glucose
lacY
part of lac operon. encodes the permease that brings lactose into the cell
lacA
part of lac operon. Encodes a transacetylase which has an unclear function in lactose metabolism
Draw out the lactose operon along with factors that bind to it in order to regulate its transcription
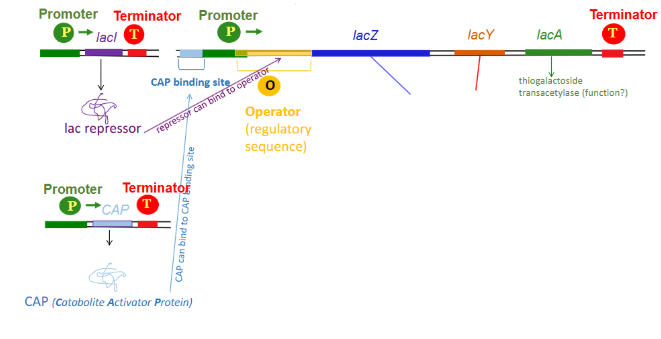
Operator
a segment of DNA where regulatory proteins, such as repressors, bind to control the transcription of nearby genes. Overlaps somewhat with the promoter in the lac operon and is where the lac repressor binds.
lacl
outside of the lac operon. Encodes the repressor for the operon which binds to the operator and blocks the sigma factor from binding
CAP
outside of the lac operon. Catabolite activator protein. Binds to CAP binding site that is upstream of the promoter to enhance transcription of the lac operon. Must be in complex with cAMP to bind to CAP-binding site (cAMP levels are only high enough for this to happen when glucose levels are low). Necessary for effiecnt transcription of lac operon
Where does the lac repressor bind?
the operator (overlapping and downstream of promoter)
Where does CAP bind (lac operon)?
CAP binding site (upstream of promoter)
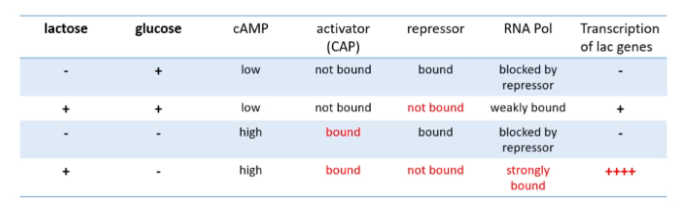
In the absence of lactose what happens with the lac operon repressor?
lac repressor protein is made via lacl gene. Lac repressor binds to lac operator which inhibits transcription
In the presence of lactose what happens with the lac operon repressor?
lac repressor is made and binds allolactose (derived from lactose) this changes the conformation of the repressor to cause it not to bind to the operator. Transcription may then occur.
If both lactose and glucose are available to a bacteria, what happens?
lac genes are not efficiently expressed as CAP needs to be in complex with cAMP to be able to bind to the CAP-binding site. CAP in this way acts as a sensor for glucose because cAMP level is high when glucose level is low.
Fill in the table
The trp operon has a —- system with two mechanisms of —- regulation
repressible; negative
Trp operon
includes 5 genes coding for enzymes that synthesis Tryptophan(Trp).
Leader sequence (Trp operon)
in 5’ UTR and contains ORF (open reading frame) which codes for a short (14 codon) peptide with 2 Trp residues
Trp repressor
made separately from the Trp operon. Represses Trp operon when Trp levels are high has it binds Trp in order to be able to bind to operator and stop transcription
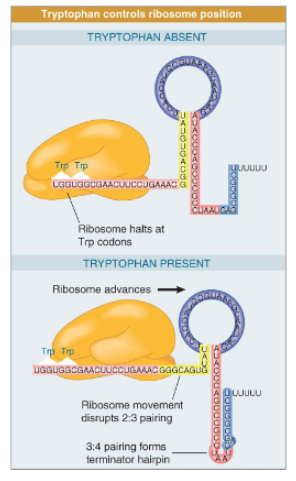
Transcriptional attenuation of Trp operon
transcription of Trp operon begins at leader and attenuator sequence. The nascent mRNA of th attenuator sequence may adopt two different structures, structure 1 which contains a terminator hairpin that causes RNA pol to release form the DNA and structure 2 which does not have the terminator hairpin. When Trp-tRNA is low, the ribosome pauses during translation of the leader mRNA as it stalls at the Trp codons which favors formation of structure 2 (allowing transcription of the Trp operon). When Trp-tRNA is high, the sequence is translated successfully which favors formation of structure 1 (inhibiting transcription of the operon).
Riboswitches
RNA that can adopt two different stable structures. common in bacteria 5’ UTR regions.may direct binding of metabolites to RNA via changes is secondary structure. May regulate transcription or translation.
Small RNA (sRNA)
~80-100 bp. Forms base pairs with mRNA (usually not perfectly). Because of this imperfect base pairing one sRNA can regulate multiple target genes. May regulate both transcription and translation.
Examples of how sRNA can control transcription/translation
may block ribosome binding, induce degradation of RNA, help the RNA to to fold in into or out of a termination loop, or provides a ribosome binding site