SBI4U- U4 Molecular Genetics: DNA, Replication, & Protein Synthesis Practice Test
1/167
Earn XP
Description and Tags
**Test Qs from 2022 l *Test Q's from 2023
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
168 Terms
1. When Fred Griffith injected mice with a mixture of dead, pathogenic, encapsulated S cells and living unencapsulated R cells of Streptococcus pneumoniae, he discovered that
a. | the previously harmless strain (S) had inherited the ability to kill mice (transformed into encapsulated cells). |
b. | the dead mice teemed with living pathogenic (R) cells. |
c. | the killer strain (R) was encased in a protective capsule. |
d. | the dead mice teemed with living pathogenic (R) cells and the killer strain (R) was encased in a protective capsule. |
e. | all of these |
d. | the dead mice teemed with living pathogenic (R) cells and the killer strain (R) was encased in a protective capsule. |
2. The significance of Fred Griffith's experiment in which he used two strains of Streptococcus pneumoniae is that
a. | the semiconservative nature of DNA replication was finally demonstrated. |
b. | it demonstrated that harmless cells had become permanently transformed through a change in the bacterial hereditary system. |
c. | it established that pure DNA extracted from disease-causing bacteria transformed harmless strains into killer strains. |
d. | it demonstrated that radioactively labeled bacteriophages transfer their DNA but not their protein coats to their host bacteria. |
e. | all of these |
b. | it demonstrated that harmless cells had become permanently transformed through a change in the bacterial hereditary system. |
3. Bacteriophages are
a. | large bacteria. |
b. | pathogens (disease-producing bacteria). |
c. | viruses. |
d. | cellular components. |
e. | protistans. |
c. | viruses. |
4. In the cycle of viral infection, which of the following is the last process to occur?
a. | invasion of viruses into the body |
b. | replication of viral particles by bacteria |
c. | lysis of the bacteria |
d. | attachment of virus to bacteria |
e. | injection of nucleic acid into the bacteria |
c. | lysis of the bacteria |
5. The significance of the experiments in which 32P and 35S were used is that
a. | the semiconservative nature of DNA replication was finally demonstrated. |
b. | it demonstrated that harmless bacterial cells had become permanently transformed through a change in the bacterial hereditary system. |
c. | it established that pure DNA extracted from disease-causing bacteria transformed harmless strains into killer strains. |
d. | it demonstrated that radioactively labeled bacteriophages transfer their DNA but not their protein coats to their host bacteria. |
e. | none of these |
d. | it demonstrated that radioactively labeled bacteriophages transfer their DNA but not their protein coats to their host bacteria. |
6. If a mixture of viruses labeled with radioactive sulfur and phosphorus is placed in a bacterial culture,
a. | the bacteria will absorb radioactive sulfur. |
b. | the bacteria will absorb radioactive phosphorus. |
c. | the bacteria will absorb both radioactive sulfur and phosphorus. |
d. | the bacteria will not absorb either sulfur or phosphorus. |
e. | the viruses will not attach to the bacteria. |
b. | the bacteria will absorb radioactive phosphorus. |
7. The Hershey-Chase experiment exploits the fact that nucleic acid contains one of the following while proteins do not. Which one?
a. | sulfur. | c. | potassium. | e. | manganese. |
b. | phosphorus. | d. | iron. |
b. | phosphorus. |
8. Sulfur is
a. | found in proteins, but not nucleic acids. |
b. | a vital component of DNA. |
c. | found in nucleic acids, but not proteins. |
d. | needed for bacteriophages to attach to bacteria. |
e. | needed for the enzyme that splits the wall of bacteria. |
a. | found in proteins, but not nucleic acids. |
9. Which of the following statements is false?
a. | Protein molecules contain no phosphorus. |
b. | Hershey and Chase discovered that 35S and not 32P had been incorporated into the hereditary system of the bacteria. |
c. | Bacteriophages are viruses that inject their nucleic acid genetic code into bacteria and use the bacterial genetic apparatus to make viral proteins. |
d. | Each nucleotide is composed of a five-carbon sugar, a phosphate group, and either a purine or pyrimidine. |
e. | Viruses are particles of nucleic acid encased in protein. |
b. | Hershey and Chase discovered that 35S and not 32P had been incorporated into the hereditary system of the bacteria. |
they discovered 35P was incorporated
10. The building blocks of nucleic acids are
a. | amino acids. |
b. | nucleotides. |
c. | pentose sugars. |
d. | phosphate groups. |
e. | nitrogenous bases. |
b. | nucleotides. |
11. Which of the following terms is NOT related to the other four?
a. | amino acids | d. | phosphate groups |
b. | nucleotides | e. | nitrogenous bases |
c. | pentose sugars |
a. | amino acids |
12. A nucleotide may contain
a. | a purine. | d. | a pyrimidine. |
b. | a pentose. | e. | All of these. |
c. | a phosphate group. |
e. | All of these. |
13. Which scientist(s) discovered the basis for the base-pair rule, which states that the amounts of adenine and thymine match, as do the amounts of cytosine and guanine?
a. | Avery | d. | Hershey and Chase |
b. | Griffith | e. | Pauling |
c. | Chargaff |
c. | Chargaff |
14. DNA varies from species to species in its
a. | base-pair bonding. |
b. | relative amounts of nucleotide bases. |
c. | sequence of base pairs. |
d. | base-pair bonding and sequence of base pairs. |
e. | relative amounts of nucleotide bases and sequence of base pairs. |
e. | relative amounts of nucleotide bases and sequence of base pairs. |
15. Rosalind Franklin's research contribution was essential in
a. | establishing the double-stranded nature of DNA. |
b. | establishing the principle of base pairing. |
c. | establishing most of the principal structural features of DNA. |
d. | sequencing DNA molecules. |
e. | determining the bonding energy of DNA molecules. |
c. | establishing most of the principal structural features of DNA. |
16. Rosalind Franklin used which technique to determine many of the physical characteristics of DNA?
a. | transformation |
b. | transmission electron microscopy |
c. | density-gradient centrifugation |
d. | x-ray diffraction |
e. | all of these |
d. | x-ray diffraction |
17. James Watson and Francis Crick
a. | established the double-stranded nature of DNA. |
b. | confirmed the principle of base pairing. |
c. | explained how DNA's structure permitted it to be replicated. |
d. | proposed the concept of the double helix. |
e. | all of these |
e. | all of these |
18. In the bonding of two nucleotides across a double helix,
a. | hydrogen bonds are used. |
b. | adenine and thymine bind together. |
c. | purines bind with pyrimidines. |
d. | double-ring nitrogenous bases connect to single-ring bases. |
e. | all of these |
e. | all of these |
19. In DNA, complementary base pairing occurs between
a. | cytosine and uracil. | d. | adenine and thymine. |
b. | adenine and guanine. | e. | all of these |
c. | adenine and uracil. |
d. | adenine and thymine. |
20. Adenine and guanine are
a. | double-ringed purines. | d. | single-ringed pyrimidines. |
b. | single-ringed purines. | e. | amino acids. |
c. | double-ringed pyrimidines. |
a. | double-ringed purines. |
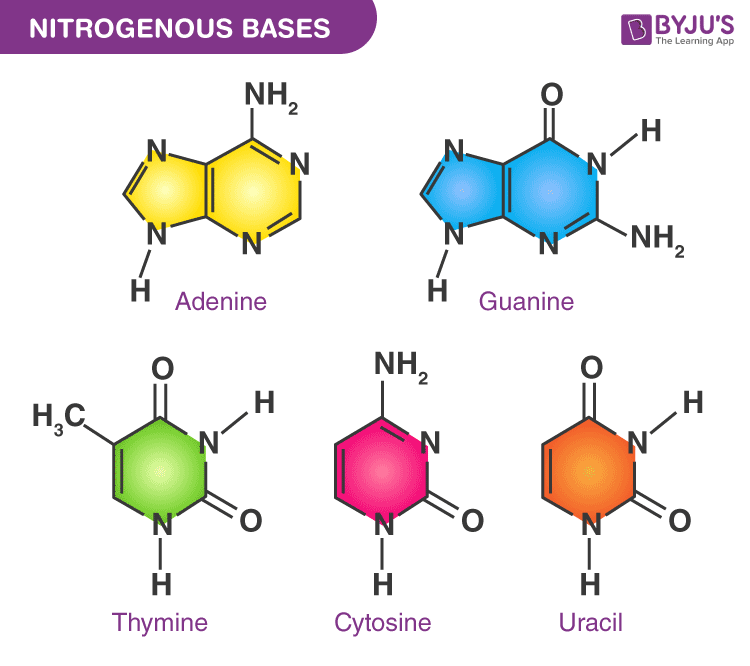
21. If a purine bonded to a purine in DNA, the molecule would __________ in that region.
a. | be constricted | d. | unwind |
b. | be perfectly normal | e. | bulge |
c. | lose a sugar-phosphate unit |
e. | bulge |
22. In the comparison between a spiral staircase and a DNA molecule, the steps would correspond to
a. | sugars. | c. | base pairs. | e. | phosphates. |
b. | hydrogen bonds. | d. | nucleotides. |
c. | base pairs. |
23. Which of the following statements is true?
a. | The hydrogen bonding of cytosine to guanine is an example of complementary base pairing. |
b. | Adenine always pairs up with guanine in DNA, and cytosine always teams up with thymine. |
c. | Each of the four nucleotides in a DNA molecule has the same nitrogen-containing base. |
d. | When adenine base pairs with thymine, they are linked by three hydrogen bonds. |
e. | In the DNA of all species, the amount of purines never equals the amount of pyrimidines. |
a. | The hydrogen bonding of cytosine to guanine is an example of complementary base pairing. |
24. Each DNA strand has a backbone that consists of alternating
a. | purines and pyrimidines. |
b. | nitrogen-containing bases. |
c. | hydrogen bonds. |
d. | sugar and phosphate molecules. |
e. | amines and purines. |
d. | sugar and phosphate molecules. |
25. The appropriate adjective to describe DNA replication is
a. | nondisruptive. | d. | natural. |
b. | semiconservative. | e. | lytic. |
c. | progressive. |
b. | semiconservative. |
26. Replication of DNA
a. | produces RNA molecules. |
b. | produces only new DNA. |
c. | produces two molecules, each of which is half-new and half-old DNA joined lengthwise to each other. |
d. | generates excessive DNA, which eventually causes the nucleus to divide. |
e. | is too complex to characterize. |
c. | produces two molecules, each of which is half-new and half-old DNA joined lengthwise to each other. |
27. DNA strands serve as which of the following during DNA synthesis?
a. | replicate | d. | source of nucleotides |
b. | substitute | e. | all of these |
c. | template |
c. | template |
28. What is the basis for the difference in how the leading and lagging strands of DNA molecules are synthesized?
a. | The origins of replication occur only at the 5' end. |
b. | Helicases and single-strand binding proteins work at the 5' end. |
c. | DNA polymerase can join new nucleotides only to the 3' end of a growing strand. |
d. | DNA ligase works only in the 3' → 5' direction. |
e. | Polymerase can work on only one strand at a time. |
c. | DNA polymerase can join new nucleotides only to the 3' end of a growing strand. |
29. Which would you expect of a eukaryotic cell lacking telomerase?
a. | a high probability of becoming cancerous |
b. | production of Okazaki fragments |
c. | inability to repair thymine dimers |
d. | a reduction in chromosome length |
e. | high sensitivity to sunlight |
d. | a reduction in chromosome length |
30. DNA polymerase
a. | is an enzyme. |
b. | adds new nucleotides to a strand. |
c. | proofreads DNA strands to see that they are correct. |
d. | derives energy from dNTP’s for synthesis of DNA strands. |
e. | all of these |
e. | all of these |
31. The leading and the lagging strands differ in that
a. | the leading strand is synthesized in the same direction as the movement of the replication fork, and the lagging strand is synthesized in the opposite direction. |
b. | the leading strand is synthesized by adding nucleotides to the 3' end of the growing strand, and the lagging strand is synthesized by adding nucleotides to the 5' end. |
c. | the lagging strand is synthesized continuously, whereas the leading strand is synthesized in short fragments that are ultimately stitched together. |
d. | the leading strand is synthesized at twice the rate of the lagging strand. |
a. | the leading strand is synthesized in the same direction as the movement of the replication fork, and the lagging strand is synthesized in the opposite direction. |
32. A linear stretch of DNA that specifies the sequence of amino acids in a polypeptide is called a(n)
a. | codon. | c. | messenger. | e. | enzyme. |
b. | intron. | d. | gene. |
d. | gene. |
33. The "central dogma" of molecular biology
a. | explains the structural complexity of genes. |
b. | describes the flow of information. |
c. | is based upon the role of proteins in controlling life. |
d. | does not explain how genes function. |
e. | explains evolution in terms of molecular biology. |
b. | describes the flow of information. |
34. All the different kinds of RNA are transcribed in the
a. | mitochondria. | c. | ribosomes. | e. | E.R. |
b. | cytoplasm. | d. | nucleus. |
d. | nucleus. |
35. The form of RNA that carries the code from the DNA to the site where the protein is assembled is called
a. | messenger RNA. | d. | transfer RNA. |
b. | nuclear RNA. | e. | structural RNA. |
c. | ribosomal RNA. |
a. | messenger RNA. |
36. The nitrogenous base found in DNA but not in RNA is
a. | adenine. | c. | guanine. | e. | thymine. |
b. | cytosine. | d. | uracil. |
e. | thymine. |
37. DNA and RNA are alike in
a. | the pentose sugar. | d. | their function in genetics. |
b. | all their nitrogenous bases. | e. | none of these. |
c. | ribosomal RNA. |
e. | none of these. |
38. Uracil will pair with
a. | ribose. | c. | cytosine. | e. | guanine. |
b. | adenine. | d. | thymine. |
b. | adenine. |
39. The synthesis of an RNA molecule from a DNA template strand is
a. | replication. | c. | transcription. | e. | metabolism. |
b. | translation. | d. | DNA synthesis. |
c. | transcription. |
40. The relationship between strands of RNA and DNA is
a. | antagonistic. | c. | complementary | e. | unrelated. |
b. | opposite. | d. | an exact duplicate. |
c. | complementary |
41. Transcription
a. | occurs on the surface of the ribosome. |
b. | is the final process in the assembly of a protein. |
c. | occurs during the synthesis of any type of RNA from a DNA template. |
d. | is catalyzed by DNA polymerase. |
e. | all of these |
c. | occurs during the synthesis of any type of RNA from a DNA template. |
42. Transcription
a. | involves both strands of DNA as templates. |
b. | uses the enzyme DNA polymerase. |
c. | results in a double-stranded end product. |
d. | produces three different types of RNA molecules. |
e. | all of these |
d. | produces three different types of RNA molecules. |
43. Transcription starts at a region of DNA called a(n)
a. | sequencer. | c. | activator. | e. | transcriber. |
b. | promoter. | d. | terminator.. |
b. | promoter. |
44. Which of the following dominates the process of transcription?
a. | RNA polymerase | c. | phenylketonuria | e. | all of these |
b. | DNA polymerase | d. | transfer RNA |
a. | RNA polymerase |
45. Before leaving the nucleus, the RNA molecule
a. | acquires a poly-A tail. |
b. | breaks loose from the terminator signal on the template. |
c. | becomes capped. |
d. | is stripped of its introns. |
e. | all of these |
e. | all of these |
46. In transcription,
a. | several RNA molecules are made from the same DNA molecule. |
b. | promoters are needed so that RNA can bind to DNA. |
c. | DNA produces messenger RNA. |
d. | a specific enzyme called RNA polymerase is required. |
e. | all of these |
e. | all of these |
47. The portion of the DNA molecule that is translated is composed of
a. | introns. | c. | exons. | e. | exons and |
b. | anticodons. | d. | transcriptons. | transcriptons. |
c. | exons. |
48. The portion of the DNA molecule that is not translated and is a noncoding portion of DNA is composed of
a. | introns. | c. | exons. | e. | exons and |
b. | anticodons. | d. | transcriptons. | transcriptons. |
a. | introns. |
49. Before messenger RNA is mature,
a. | all exons are deleted and removed. |
b. | a cap and a tail are provided. |
c. | anticodons are assembled. |
d. | the transfer RNA transfers the messenger RNA to the ribosome. |
e. | the single RNA strand duplicates itself in much the same way as DNA. |
b. | a cap and a tail are provided. |
50. If the DNA triplets were ATG-CGT, the mRNA codons would be
a. | AUG-CGU. | c. | UAC-GCA. | e. | none of these |
b. | ATG-CGT. | d. | UAG-CGU. |
c. | UAC-GCA. |
51. If the DNA triplets were ATG-CGT, the tRNA anticodons would be
a. | AUG-CGU. | c. | UAC-GCA. | e. | none of these |
b. | ATG-CGT. | d. | UAG-CGU. |
a. | AUG-CGU. |
transcription: mRNA- UAC-GCA
translation: complementary tRNA anti codons- AUG-CGU
52. In transcription,
a. | several amino acids are assembled by the messenger RNA molecules at one time. |
b. | a special sequence called a promoter is necessary for transcription to begin. |
c. | certain polypeptide sequences are governed by one ribosome, whereas other sequences are produced by other ribosomes. |
d. | the transfer RNA molecules arrange the messenger RNA codons into the appropriate sequence. |
e. | none of these |
b. | a special sequence called a promoter is necessary for transcription to begin. |
53. The genetic code
a. | is universal for all organisms. |
b. | is based upon 64 codons made of sequences of three nucleotides. |
c. | also comes equipped with punctuation marks. |
d. | is redundant, that is, each amino has more than one codon. |
e. | all of these |
e. | all of these |
If the codon consisted of only two nucleotides, there would be how many different kinds of codons?
a. 4
b. 8
c. 16
d. 32
e. 64
c. 16
# of codons= (# of possible bases)length of codon
Since there are 4 possible bases (A, U, G, C for RNA), and each codon consists of 2 nucleotides:
= 42
=16
The insertion of how many nucleotides into a genetic sequence does less damage to the code than the insertion of other numbers of nucleotides?
a. 1
b. 2
c. 3
d. 4
e. 5
c. 3
56. The concept that a set of three nucleotides specifies a particular amino acid provides the basis for
a. | the one gene, one enzyme hypothesis. |
b. | the one gene, one polypeptide hypothesis. |
c. | the genetic code. |
d. | biochemical reactions among nucleic acids. |
e. | all of these |
c. | the genetic code. |
57. Of all the different codons that exist, three of them
a. | are involved in mutations. |
b. | do not specify a particular amino acid. |
c. | cannot be copied. |
d. | provide punctuation or instructions such as "stop." |
e. | do not specify a particular amino acid and provide punctuation or instructions such as "stop." |
e. | do not specify a particular amino acid and provide punctuation or instructions such as "stop." |
58. Crick and Brenner discovered that the presence of three extra nucleotides inserted in the middle of a gene caused far fewer problems than if only one or two extra nucleotides were inserted. They interpreted this result to mean that
a. | the genetic code consists of nonoverlapping triplets of nucleotide bases. |
b. | the longer the sequence of nucleotides that is added to a gene, the more chemically stable the resulting DNA is. |
c. | there had been significant experimental error in their electrophoresis studies. |
d. | the wobble effect accounts for the unpredictability in codon-anticodon pairing at the third base. |
e. | all of these |
a. | the genetic code consists of nonoverlapping triplets of nucleotide bases. |
59. The wobble effect
a. | explains why and how there can be 31 kinds of transfer RNA molecules. |
b. | allows the third codon to vary if the first two codons in the anticodon follow the base-pair rule. |
c. | indicates that transfer RNA combines with either the small or large subunit of ribosomes. |
d. | explains why and how there can be 31 kinds of transfer RNA molecules, and allows the third codon to vary if the first two codons in the anticodon follow the base-pair rule. |
e. | explains why and how there can be 31 kinds of transfer RNA molecules, allows the third codon to vary if the first two codons in the anticodon follow the base-pair rule, and indicates that transfer RNA combines with either the small or large subunit of ribosomes. |
d. | explains why and how there can be 31 kinds of transfer RNA molecules, and allows the third codon to vary if the first two codons in the anticodon follow the base-pair rule. |
60. Each "word" in the mRNA language consists of how many letters?
a. | 3 | c. | 5 | e. | none of these |
b. | 4 | d. | more than 5. |
a. | 3 |
61. Transfer RNA differs from other types of RNA because it
a. | transfers genetic instructions from cell nucleus to cytoplasm. |
b. | specifies the amino acid sequence of a particular protein. |
c. | carries an amino acid at one end. |
d. | contains codons. |
e. | none of these |
c. | carries an amino acid at one end. |
62. The wobble effect pertains to the matching of
a. | codons with anticodons. |
b. | codons with exons. |
c. | exons with introns. |
d. | template DNA with messenger RNA. |
e. | messenger RNA with ribosomal RNA. |
a. | codons with anticodons. |
63. Eukaryotic ribosomes function as
a. | a single unit. | d. | four-part units. |
b. | two-part units. | e. | a multidivisional unit. |
c. | three-part units. |
b. | two-part units. |
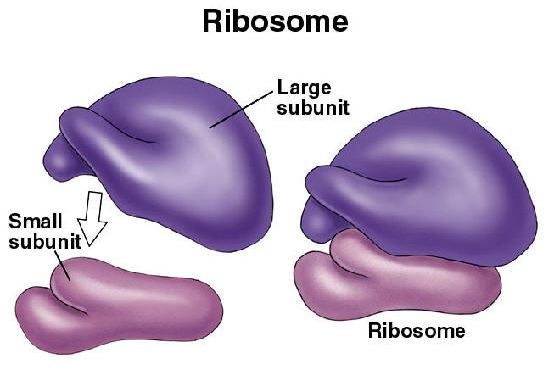
64. All mRNA transcripts begin with
a. | methionine. | c. | AUG. | e. | anticodon. |
b. | a ribosome. | d. | the P site. |
c. | AUG. |
65. Which of the following statements is FALSE?
a. | In chain elongation, the amino acids are added to the chain according to the sequence in the messenger RNA. |
b. | The messenger RNA molecule is stationary and series of ribosomes called polysomes travel along the molecule manufacturing series of polypeptides at the same time. |
c. | The shape of transfer RNA molecules is uniform and is maintained by hydrogen bonds. |
d. | Enzymes found in the ribosome catalyze the formation of the bonds in the new polypeptide. |
e. | None of these statements is false. |
c. | The shape of transfer RNA molecules is uniform and is maintained by hydrogen bonds. |
66. A polysome is
a. | one of the units of a ribosome. |
b. | the nuclear organelle that synthesizes RNA. |
c. | an organelle that functions similarly to a ribosome during meiosis. |
d. | the two units of a ribosome considered together. |
e. | an mRNA molecule with several ribosomes attached. |
e. | an mRNA molecule with several ribosomes attached. |
67. A gene mutation
a. | is a change in the nucleotide sequence of DNA. |
b. | may be caused by environmental agents. |
c. | may arise spontaneously. |
d. | can occur in any organism. |
e. | all of these |
e. | all of these |
68. Which of the following statements is true?
a. | Gene mutations occur independently of each other. |
b. | Gene mutations are relatively rare. |
c. | The reason that two antibiotics may be given at one time is due to the very remote possibility that two gene mutations could occur in the same cell at the same time. |
d. | Mutations are random; that is, it is impossible to predict exactly when a specific gene will mutate but an expected frequency can be assigned. |
e. | all of these |
e. | all of these |
69. Mutations can be
a. | random. | c. | lethal. | e. | All of these. |
b. | beneficial. | d. | heritable. |
e. | All of these. |
70. Frameshift mutations may involve
a. | substitution of nucleotides. |
b. | substitution of codons. |
c. | substitution of amino acids. |
d. | addition or deletion of one to several base pairs. |
e. | all of these |
d. | addition or deletion of one to several base pairs. |
71. Sickle-cell anemia has been traced to what type of mutation?
a. | frameshift | d. | base-pair substitution |
b. | transposable element | e. | viral |
c. | mutagenic |
d. | base-pair substitution |
72. The difference between normal and sickle-cell hemoglobin is based upon
a. | the number of amino acids in the molecule. |
b. | the substitution of one amino acid for another. |
c. | the number and orientation of the amino acid chains attached to the heme portion of the molecule. |
d. | the number of oxygen molecules that can be carried. |
e. | the type of bone marrow that produces it. |
b. | the substitution of one amino acid for another. |
73. In a mutation,
a. | the new codon may specify a different amino acid, but may not change the function of the new protein produced. |
b. | the new codon may specify the same amino acid as the old codon. |
c. | the new codon and resulting amino acid may destroy the function of the protein specified. |
d. | All of these may be true. |
d. | All of these may be true. |
74. At a certain location in a gene, the nontemplate strand of DNA has the sequence GAA. A mutation alters the triplet to GAG. This type of mutation is called
a. | silent. | c. | nonsense. | e. | translocation. |
b. | missense. | d. | frame-shift. |
a. | silent. |
75. A mutation that results in the codon UAG where there had been UCG is
a. | a nonsense mutation. |
b. | a missense mutation. |
c. | a frame-shift mutation. |
d. | a large-scale mutation. |
e. | unlikely to have a significant effect. |
a. | a nonsense mutation. |
76. Four of the five answers listed below are steps in the process of transcription. Select the exception.
a. | cap put on one end | d. | poly-A tail at one end |
b. | introns snipped out | e. | exons spliced together |
c. | action by DNA polymerase |
c. | action by DNA polymerase |
77. Four of the five answers listed below are related pairings. Select the exception.
a. | double-stranded DNA - mRNA |
b. | purine - pyrimidine |
c. | codon - anticodon |
d. | small subunit - large subunit |
e. | promoter - terminator |
e. | promoter - terminator |
78. Four of the five answers listed below describe changes at the chromosomal level. Select the exception.
a. | base substitution | c. | translocation | e. | inversion |
b. | duplication | d. | deletion |
a. | base substitution |
79. Four of the five answers listed below are components of a nucleotide. Select the exception.
a. | pentose sugar | d. | phosphate group |
b. | amino acid | e. | purine |
c. | pyrimidine |
b. | amino acid |
80. Three of the four answers listed below are steps in translation. Select the exception.
a. | initiation | c. | chain elongation | e. | inversion |
b. | replication | d. | termination |
b. | replication |
Matching Match each item with the correct statement below.
a. | template strand | i. | single-stranded binding proteins |
b. | ligase | j. | leading strand |
c. | primase | k. | gyrase |
d. | DNA polymerase I | l. | helicase |
e. | lagging strand | m. | annealing |
f. | replication fork | n. | mitosis |
g. | semi-conservative replication | o. | DNA polymerase III |
h. | Okazaki fragment | ||
81. process which results in each new DNA molecule consisting of one parental strand and one newly synthesized strand
82. unwinds DNA
83. removes RNA primers
84. new DNA strand synthesized in fragments
85. division of a nucleus to form two daughter nuclei
86. short lengths of DNA produced during synthesis of lagging strand
87. keep newly separated strands of DNA apart
88. new DNA strand which is synthesized continuously
89. links or repairs gaps in the sugar-phosphate backbone
90. area where DNA polymerase is bound to unwound DNA
91. DNA strand which directs synthesis of a complementary strand
92. builds RNA primers
93. relieves tension in DNA during unwinding
94. pairing of complementary strands of DNA due to hydrogen bonding
95. responsible for building new DNA strands during replication
G
L
D
E
N
H
I
J
B
F
A
C
K
M
O
For the following groups, the same choice may be used more than once as an answer, or not at all.
Questions 96-99 refer to the following enzymes
a. DNA Ligase
b. DNA polymerase
C. RNA polymerease
D. Restriction renzyme
E. Reverse transcriptase
Enzyme used in the synthesis of mRNA
Enzyme used during replication to attach Okazaki fragments to each other
Enzyme found in retro viruses that produce DNA from an RNA template
Enzyme used to position nucleotides during DNA replication
C
A
E
B
Questions 100-103 refer to the following groups of biological compounds
(A) proteins
(B) carbohydrates
(C) nucleic acids
(D) lipids
(E) steroids
100. Synthesized at the ribosome.
101. Includes glycogen, chitin, cellulose, and glucose.
102. Used for insulation and buoyancy in marine Arctic animals
103. Used to carry the genetic code.
A
B
D
C
Questions 104-106 refer to the following elements of the synthesis of a polypeptide on a ribosome.
(A) Binding of an amino acid to tRNA.
(B) Translocation of the ribosome along the mRNA strand.
(C) Separation of large and small ribosomal subunits.
(D) Binding of tRNA to mRNA.
(E) Involves codon-anticodon recognition.
104. Involves the formation of a peptide bond.
105. Triggered by stop codon.
106. Involves codon-anticodon recognition.
E
C
D
*For DNA, the base pairing rules first proposed by (i) ______, tell us that if 23% of the bases are adenine, then (ii) _______ of the bases must be cytosine
a. Avery; 23%
b. Avery; 27&
c. Chargaff; 23%
d. Chargaff; 27%
e. Crick; 23%
f. Crick 27%
d. Chargaff; 27%

*According to Beadle and Tatum’s hypothesis, how many genes are necessary for the following pathway
a. 0
b. 1
c. 2
d. 3
d. 3
*Which enzyme catalyzes the elongation of a DNA strand in the 5’ to 3’ direction
a. primase
b. DNA ligase
c. DNA polymerase III
d. topoisomerase
e. helicase
c. DNA polymerase III
*Which of the following removes the RNA nucleotides from the primer and adds equivalent DNA nucleotides to the 3’ end of Okazaki fragments?
a. helicase
b. DNA pol III
c. ligase
d. DNA pol I
e. primase
d. DNA pol I
*The primary function of DNA ligase is to
a. cut the two strands of the DNA molecule prior to replication
b. attach free nucleotides to the growing chain
c. remove bases that might have been inserted incorrectly
d. seal new short stretches into one continuous strand
e. fragment old DNA that is no longer of use to the cell
d. seal new short stretches into one continuous strand
*RNA polymerase moves in which direction along the DNA?
a. 3’ to 5’ along the template strand
b. 3’ to 5’ along the coding (sense) strand
c. 5’ to 3’ along the template strand
d. 3’ to 5’ along the coding strand
e. 5’ to 3’ along the double stranded DNA
a. 3’ to 5’ along the template strand
think… if RNApol only builds 5’-3’, then the template strand is the one running 3’-5’ from the start point
*Introns are significant to biological evolution because
a. their presence allows exons to be shuffled
b. they protect mRNA from degeneration
c. they are translated into essential amino acids
d. they maintain the genetic code by preventing incorrect DNA base pairings
e. they correct enzymatic alterations of DNA bases
a. their presence allows exons to be shuffled
Introns (non-coding regions) can play a role in evolution because they enable alternative splicing and exon shuffling, leading to the creation of new proteins and potentially increasing the diversity of proteins that an organism can produce. This can drive evolutionary innovation and adaptation.
*Which one of the following is NOT a function of tRNA
a. picking up the amino acid specified by the anticodon
b. recognizing the appropriate codons in mRNA
c. transferring nucleotides to rRNA
d. translating codons into amino acids
c. transferring nucleotides to rRNA
*as a ribosome translocates along an mRNA molecule by one codon, which of the following occurs?
a. the tRNA that was in the A site moves into the P site
b. the tRNA that was in the P site moves into the A site
c. the tRNA that was in the A site moves to the E site and is released
d. the tRNA that was in the A site departs from the ribosome via a tunnel
e. the polypeptide enters the E site
a. the tRNA that was in the A site moves into the P site
*choose the answer that has these events of protein synthesis in the proper sequence
the anticodon/codon matching is checked
the ribosome catalyzes a peptide bond formation between the neew a.a and the growing polypeptide chain
the uncharged tRNA leaves through the E site
an aminoacyl-tRNA binds to the A site
the ribosome translocates the transcript one full codon
a. 1,3,2,4,5
b. 4,1,2,5,3,
c. 5,4,3,2,1
d. 4,1,3,2,5
e. 2,4,5,1,3
b. 4,1,2,5,3
An aminoacyl-tRNA binds to the A site.
The anticodon/codon matching is checked.
The ribosome catalyzes a peptide bond formation between the new amino acid and the growing polypeptide chain.
The ribosome translocates the transcript one full codon.
The uncharged tRNA leaves through the E site.
*mRNA codons specify an amino acid, but only 45 tRNAs exist. this is best explained by the fact that…
a. some tRNAs have anti codons that recognize four or more different codons
b. the rules for base pairings between the third base of a codon an tRNA are flexible
c. many codons are never used, so the tRNA s that recognize them are dispensable
d. the DNA codes for all 61 tRNAS but some are then destroyed
e. competitive exclusion forces some tRNAs to bee destroyed by nucleases
b. the rules for base pairings between the third base of a codon an tRNA are flexible
wobble!
**The Hershey-Chase experiment exploits the fact that nucleic acid contains one of the following while proteins do not. Which one of the following is NOT found in great amounts in proteins?
a. sulfur
b. phosphorus
c. potassium
d. iron
e. manganese
b. phosphorus
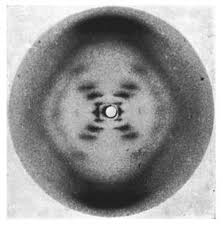
**2. Rosalind Franklin produced the famous Photo 51, used to determine many of the physical characteristics of DNA using which experimental technique?
a. transformation
b. transmission electron microscopy
c. density-gradient centrifugation
d. x-ray diffraction
e. radiolabeling
d. x-ray diffraction
**For DNA, the base-pairing rules first proposed by (i) _______, tell us that if the 27% of the bases are adenine, then (ii) _______ of the bases must be cytosine.
a. Avery; 21%
b. Avery; 27%
c. Chargaff; 21%
d. Chargaff; 27%
e. Crick; 21%
f. Crick; 27%
c. Chargaff; 21%
should be 23% not 21%…
27% A, therefore 27%T
leaves 46% left and when split into two, 23% each of G and C
**According to Beadle and Tatum's hypothesis, how many genes are necessary for the following pathway?
A enzyme A → B enzyme B → C
a. 0
b. 1
c. 2
d. 3
c. 2
**James Watson and Francis Crick
a. established the double-stranded nature of DNA.
b. confirmed the principle of base pairing.
c. explained how DNA's structure permitted it to be replicated.
d. proposed the concept of the double helix.
e. All of these.
e. All of these.