BIO 340 Exam 3
1/249
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
250 Terms
1 nitrogenous base, 1 5-carbon sugar, and 1 (or more) phosphate groups
What is the composition of a nucleotide?
RNA has one extra oxygen on it’s 5-carbon sugar
What is the main difference between RNA and DNA?
nucleosides don’t have an extra phosphate
what is the difference between a nucleoside and nucleotide?
A, D, E
Select the nitrogenous bases that are pyrimidines
A. Thymine
B.Adenine
C.Guanine
D.Uracil
E.Cytosine
B, C
Select the nitrogenous bases that are purines
A. Thymine
B.Adenine
C.Guanine
D.Uracil
E.Cytosine
C
Which is the basic pairing of nitrogenous bases?
A. Purine and Purine
B. Pyrimidine and Pyrimidine
C. Purine and Pyrimidine
pyrimidines
Nitrogenous base that is a aromatic (ring) heterocyclic. There is a nitrogen at carbons 1 and 3
purine
Nitrogenous base that is an aromatic (ring) heterocyclic plus imidazole (additional ring)
it is missing an oxygen in it’s carbon ring
This is Deoxyguanosine 5’ -monophosphate (dGMP), or the guanine nucleotide base. What does the “deoxy,” or “d,” tell us about the molecule?
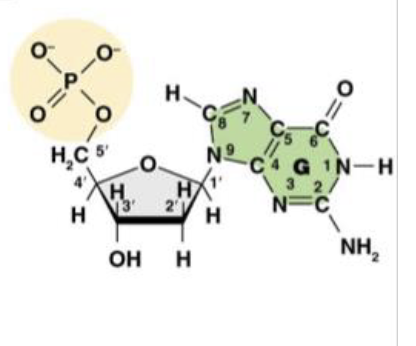
it is the nitro base present in the molecule
This is Deoxyguanosine 5’ -monophosphate (dGMP), or the guanine nucleotide base. What does the “guanosine,” or “G,” tell us about the molecule?
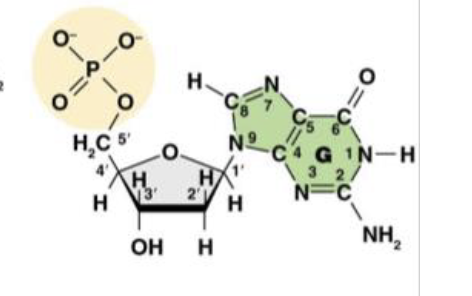
There is only one phosphate group in the molecule
This is Deoxyguanosine 5’ -monophosphate (dGMP), or the guanine nucleotide base. What does the “monophosphate,” or “MP,” tell us about the molecule?
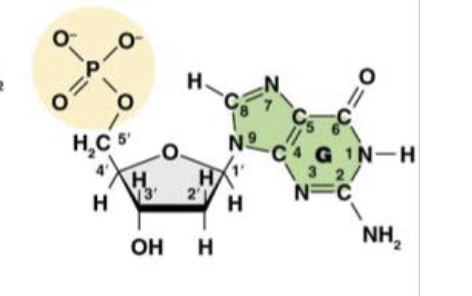
1’ is the carbon attached to the nitrogenous base
This is the molecule dGMP. What is the purpose of the 1’ carbon?
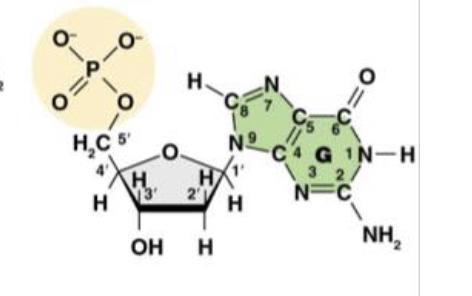
2’ is the “deoxy” carbon where only hydrogen attatches
This is the molecule dGMP. What is the purpose of the 2’ carbon?
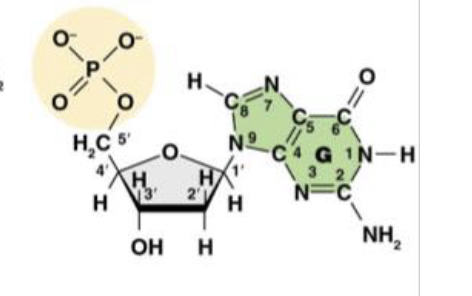
3’ the the carbon that carries the hydroxyl group where the phosphate group of another nucleotide can attach
This is the molecule dGMP. What is the purpose of the 3’ carbon?
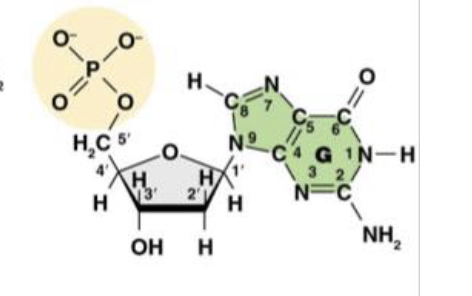
clockwise
When numbering carbons of the sugar in DNA, you should number (clockwise/counterclockwise)
it is the carbon that binds to O
This is the molecule dGMP. What is the purpose of the 4’ carbon?
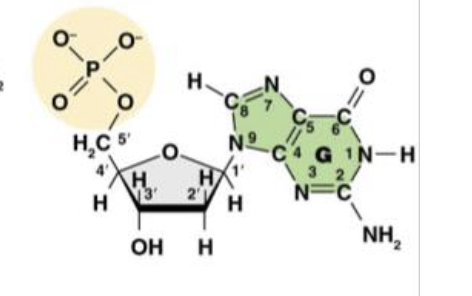
It carries the phosphate group
This is the molecule dGMP. What is the purpose of the 5’ carbon?
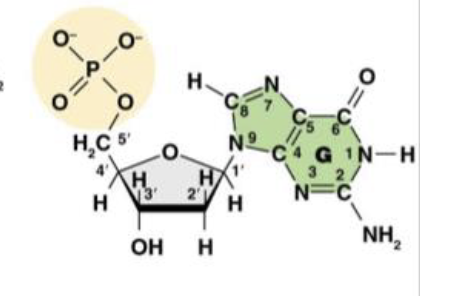
3’ and 5’
Which carbons on the carbon rings are the most important
The phosphate group that the 5’ carbon is attracted to and the hydroxyl group of the 3’ carbon form a phosphodiester bond. This links nucleotides to create the DNA backbone
Why are the 3’ and 5’ carbons of the 5-carbon ring so important?
polarity
DNA strands have ______, meaning each strand has a 5’ end and a 3’ end
Look at the 5’ carbon ring’s direction
Here is a DNA strand. What is one easy way to tell what is the polarity of the strand?
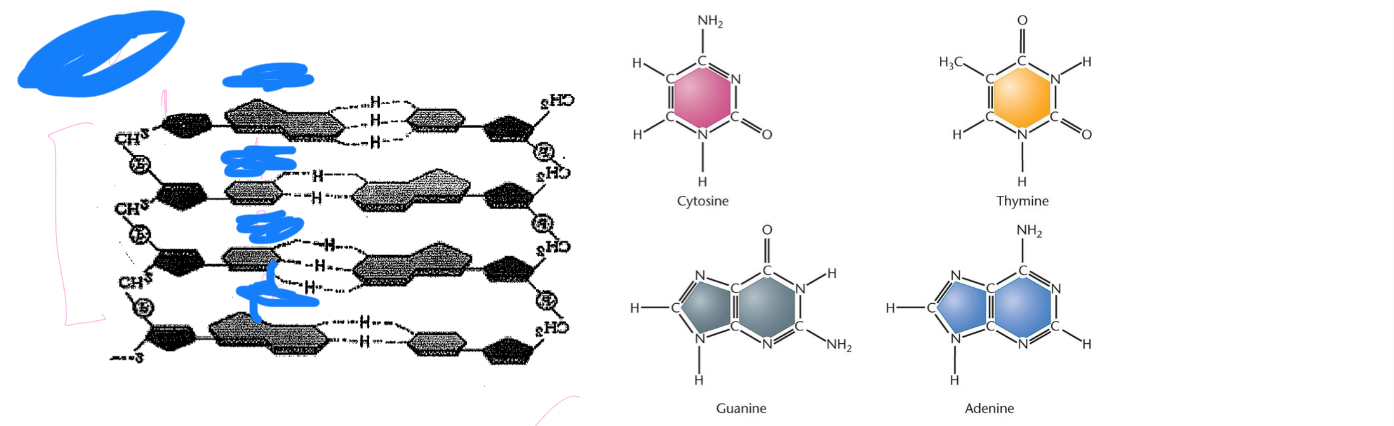
5’ to 3’
What is the polarity of this DNA strand?
C
Bases pair by what type of bonding?
A. Covalent
B. Ionic
C. Hydrogen
A
Nucleotide pair by what type of bonding?
A. Covalent
B. Ionic
C. Hydrogen
DNA strands are (parallel/antiparallel) where one strand is the (reverse/direct) complement of the other
antiparallel, reverse
thymine and adenine
Which base pair bonds are weaker: thymine & adenine or cytosine & guanine
adenine and thymine
which nucleotide base pair has two hydrogen bonds?
cytosine and guanine
which nucleotide base pair has three hydrogen bonds?
A
Adenine pairs with:
A. Thymine
B. Cytosine
C: Guanine
D: Adenine
D
Thymine pairs with:
A. Thymine
B. Cytosine
C: Guanine
D: Adenine
B
Guanine pairs with:
A. Thymine
B. Cytosine
C: Guanine
D: Adenine
C
Cytosine pairs with:
A. Thymine
B. Cytosine
C: Guanine
D: Adenine
C
Four DNA molecules are gradually heated. A strand from each are listed down below. Which molecule will melt (= denature) last?
A. ATAACGATTA
B. ACGGATGCAC
C. GCGCCCGGG
D. CGCCGGATG
3’ TTACCGAAT 5’
A portion of one strand of DNA has the sequence
5’ AATGGCTTA 3’
What is the sequence of the other strand in the 3’ to 5’ direction?
5’ TAAGCCATT 3’
A portion of one strand of DNA has the sequence
5’ AATGGCTTA 3’
What is the sequence of the other strand in the 5’ to 3’ direction?
Chargaff’s Rule
Double-stranded DNA from any cell of all organisms should have a 1:1 ratio of pyrimidine and purine bases. There fore, the amount of guanine is equal to cytosine and the amount of adenine is equal to thymine
false
True or false: the amount of A+T in a double-strands is equal to the amount of C+G
B
A sample of double-stranded DNA was found to contain 17% guanine nucleotides. Based on this, what is the percentage of adenine in the sample?
A. 17%
B. 33%
C. 34%
D. 66%
E. It depends on the sequence
C
You analyze the nucleotide content of sample DNA. Which of the results below is most consistent with the DNA being single-stranded DNA rather than double-stranded DNA?
A. G=30% A=20% C=30% T=20%
B. G=40% A=10% C=40% T=10%
C. G=30% A=30% C=20% T=20%
D. G=25% A=25% C=25% T=25%
major groove
Base functional groups are exposed on the outside of the double strand. This allows sequence to be recognized even in closed, double-stranded DNA. Proteins can also bind to this end
minor groove
Not as many functional groups exposed, but still “readable” on the outside. Less likely to have proteins bind to the DNA
True
True or false: DNA is not completely symmetrical
semiconservative
DNA is ____, meaning each daughter molecule is half old and half new, with the old serving as the template strand
C
You are observing the process of DNA replication. A free nucleotide is about to attach to the new DNA strand. How many phosphates are on the free nucleotide?
A. 1
B. 2
C. 3
D. 0
A
You are observing the process of DNA replication. A free nucleotide attaches to the new DNA strand. How many phosphates are on the nucleotide?
A. 1
B. 2
C. 3
D. 0
B
You are observing the process of DNA replication. A free nucleotide is about to attach to the new DNA strand. How many phosphates detach from the nucleotide, giving it the energy to bind to the hydroxyl group?
A. 1
B. 2
C. 3
D. 0
Before, DNA polymerase
Base pairing must happen (before/after) the phosphodiester bond can be formed. This is catalyzed by __________
5’ to 3’
DNA polymerase can only synthesize nucleotides in the ___________ direction
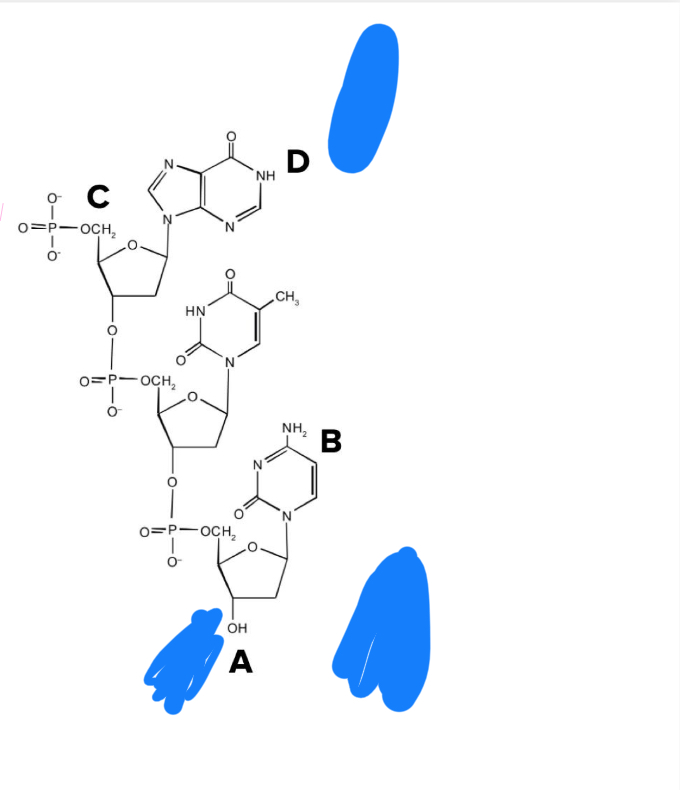
A
What sequence of the left strand of DNA pictured?
A. 5’-ACTG-3’
B. 5’-GTCA-3’
C. 5’-TAGT-3’
D. 5’-TGAT-3’
1
Pictured is a portion of a DNA daughter strand in the process of replication. Its complementary template strand is not shown. Where will the next DNA nucleotide be covalently attached?
A
B
C
D
3’ to 5’, same
During DNA synthesis, the template strand for the leading strand is going from ________ and in the (same/different) direction the parent DNA separates
5’ to 3’, different
During DNA synthesis, the template strand for the lagging strand is going from ________ and in the (same/different) direction the parent DNA separates
Okazaki fragments
The small segments of DNA that are replicated on the lagging strands
Bidirectional
DNA replication is _______, meaning two replication forks travel in two different directions on a chromosome scale
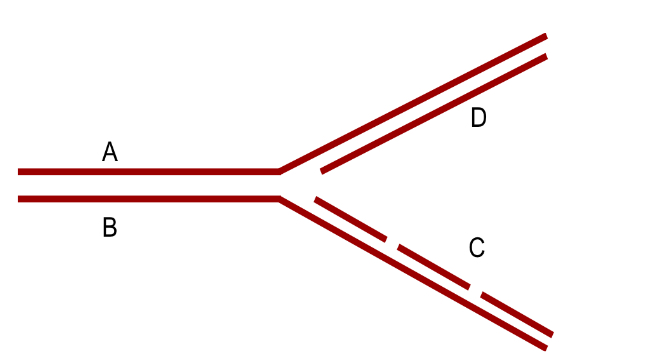
B
Which are the leading strands in this replication bubble?
A. X & Y
B. X & Z
C. X & W
D. Y & W
E. Y & Z
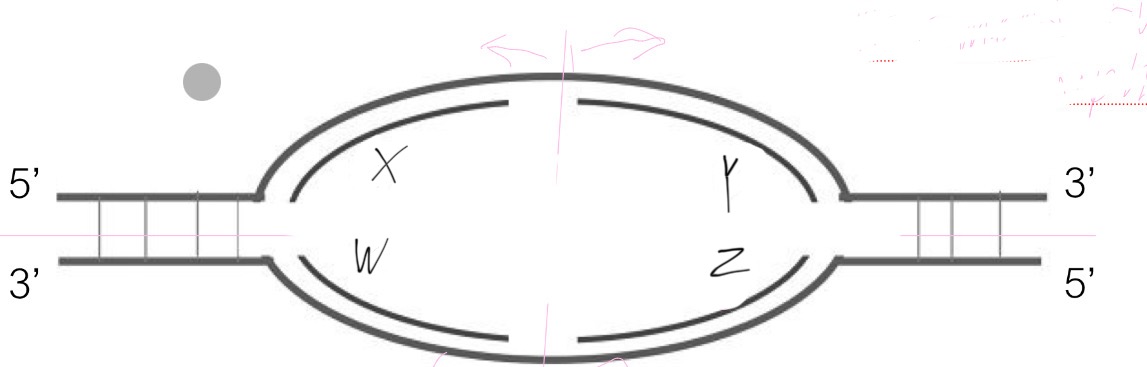
D
Which are the lagging strands in this replication bubble?
A. X & Y
B. X & Z
C. X & W
D. Y & W
E. Y & Z
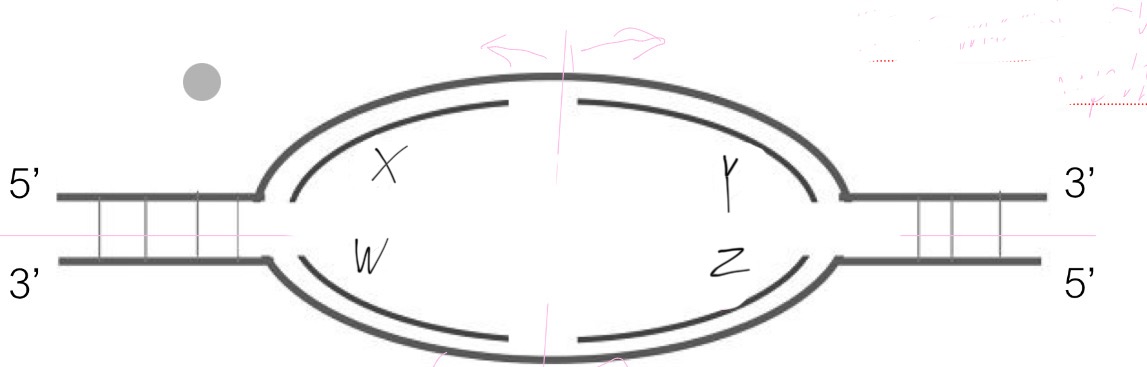
Strand 1
At Fork 1, which template strand (1 or 2) is copied in a continuous manner?
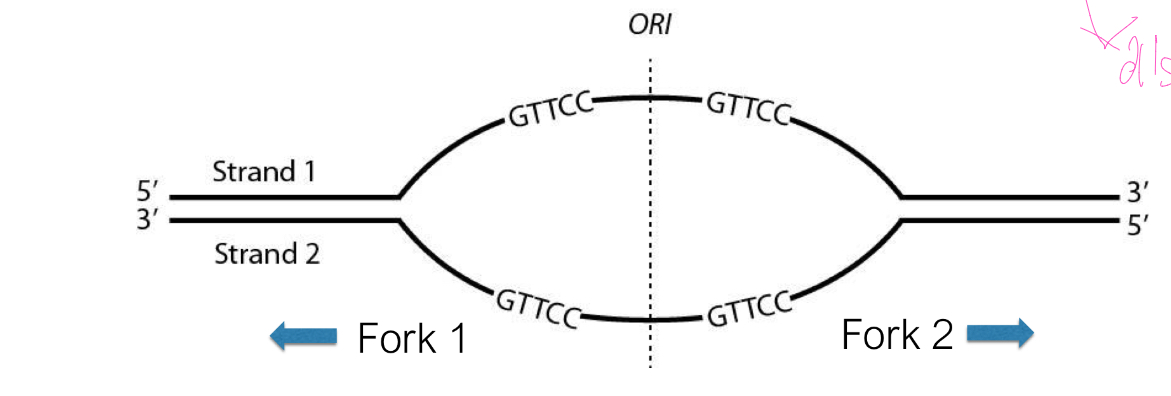
E
To which sites can the primer 5’-CAAGG-3’ bind to initiate replication
A. All sites
B. W and X
C. W and Y
D. W and Z
E. Y and Z
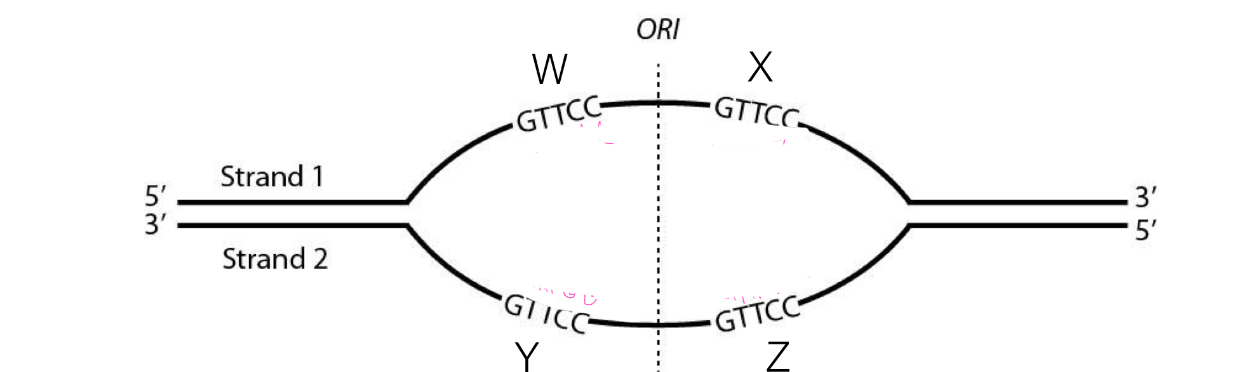
B, C
Select all the reasons why DNA replication involves a leading strand and a lagging strand
A. DNA is semiconservative
B. DNA is anti-parallel
C. DNA is bi-directional
D. DNA polymerase can only add nucleotides in the 5’ to 3’ direction
E. All of the above
False
True or false: at all levels of DNA replication is Bidirectional
Prokaryote: A, D, F, I, J
Eukaryote: B, C, E, G, H
Categorize these between prokaryote and eukaryote chromosome structure and DNA replication:
A. 1 ORI
B. Wound around histones
C. Linear chromosomes
D. Smaller chromosomes, so DNA polymerase can travel all the way around
E. Multiple ORIs
F. Free in the cytoplasm
G. Enclosed in the nucleus
H. Larger chromosomes, so one DNA polymerase cannot travel all the way around
I. Naked, no histones
J. Circular chromosomes
DNA polymerase III
Synthesizes DNA in the 5’ to 3’ direction, has 3’ to 5’ exonuclease activity, requires double stranded DNA or DNA-RNA hybrid to bind so it can extend one strand, and does the bulk of copying during DNA replication
3’ to 5’ exonuclease activity
When DNA pol III recognizes a mutation, the DNA strand slips out of its palm to another active sight where it is broken off of the DNA
Primase
Synthesizes a short stretch of RNA to make a double-stranded region that DNA pol III can bind to. Also categorized as a form of RNA polymerase
DNA polymerase I
Removes RNA nucleotides and replaces them with DNA nucleotides. Mainly used for replacing primers. I has 5’ to 3’ exonuclease activity as well as 5’ to 3’ polymerase activity
DNA ligase
Seals the “nick” (single strand break) left by the action of DNA polymerase I. This requires ATP.
Helicase
Separated DNA strands, opening the helix (by breaking hydrogen bonds) works with binding proteins (DnaA) and loading enzyme (DnaC). Also known as DnaB in Ecoli
Single strand binding proteins
Prevents re-annexing of the DNA strands. Also known as SSBs
Topoisomerase
Relaxes supercoiling by cutting the DNA, allowing it to uncoil, then re-sealing the break
4 2 1 3
Order these in the basic process of DNA replication
Replication fork proceed away from the origin in both directions making a replication bubble
All enzymes join in
Replication is continuous on the leading strand and discontinuous on the lagging strand
Double-stranded DNA separated at the origin of replication
The 13- mer will re-anneal
What would happen if SSBs were not present for DNA replication? (Hint: think about initiation)
initiation
DnaA proteins bind to 9-mer repeat sequences, bending the DNA strand and causing the AT-rich 13-mer repeats to pull apart (=melt =denature). SSBs prevent strand from re-annealing
DnaA proteins would have noting to bind to
What would happen if 9-mer wasn’t present from DNA replication? (Hint: think of initiation)
After DnaA attaches to the DNA, nothing will cause the DNA to pull apart
What would happen if 13-mer wasn’t present ? (Hint: think of initiation)
Nothing would initiate the DNA strands to break apart for replication
What would happen if DnaA wasn’t present ? (Hint: think of initiation)
OriC
Typically a few hundred bass pairs long with 9-mer (where DnaA binds to) and 13-mer (AT-rich portion that denatures quickly). Usually the one origin or replication in prokaryotes
ARSs
Origins or replications in yeast. Less conserved and do not all participate in replication during every cell cycle
Second step in DNA replication
Helicase loading protein (DnaC) loads the Helicase enzyme (DnaB) onto each single strand where it can begin separating DNA strands
Helicase wouldn’t be able to bind to DNA, thus not being able to separate the strands
What would happen if DNA Helicase loader wasn’t present for DNA replication? (Hint: think of step 2)
The DNA strands would not be continuously separated
What would happen if Helicase wasn’t present for DNA replication? (Hint: think of step 2)
Step 3 of DNA replication
Primase synthesizes the first RNA primers
DNA polymerase would have nothing to bind onto to start DNA replication
What would happen if primase wasn’t present for DNA replication? (Hint: think of step 3)
Step 4 of DNA replication
DNA polymerase III enzymes are recruited to each replication fork. DNA polymerase lll commences replication from each primer in the 5’ to 3’ direction
DNA wouldn’t be replicated
What would happen if DNA polymerase III wasn’t present for DNA replication? (Hint: think of step 4)
Step 5 of DNA replication
DNA polymerase II continues replication continuously on the leading strand and discontinuously on the lagging strand
D
During DNA replication, which enzyme is responsible for building the new DNA strands?
A. Helicase
B. Primase
C. DNA polymerase I
D. DNA polymerase III
E. ligase
A
Durning DNA replication, which enzyme is responsible for unwinding the strands of the parental DNA
A. Helicase
B. Primase
C. DNA polymerase I
D. DNA polymerase III
E. ligase
B
During DNA replication, which enzyme is responsible for adding the RNA primer?
A. Helicase
B. Primase
C. DNA polymerase I
D. DNA polymerase III
E. ligase
D
During DNA replication, which enzyme is responsible for proofreading?
A. Helicase
B. Primase
C. DNA polymerase I
D. DNA polymerase III
E. ligase
C
During DNA replication, which enzyme is responsible for removing the RNA primers?
A. Helicase
B. Primase
C. DNA polymerase I
D. DNA polymerase III
E. ligase
Nick, double-stranded
There are two ways that topoimerases help with supercoiling: ____ which is a single-stranded break, and where it makes a _____ break, passes the loop through, and then seals the gap
Telomerase
Ribonucleoprotein that has an RNA component that matches with the telomere repeat sequences at the end of DNA so it can extend the parent strand and has a protein component that uses the RNA component to synthesize DNA. Takes place on the lagging strand
The ends of chromosomes would degrade more quickly
What would happen if Telomerase didn’t exist?
B
Pictured is a representation of a replication fork, what the leading strand on top and the lagging strand on the bottom. Each red line is a single stranded DNA molecule. Where two lines run parallel, there are complementary base pairings to form double stranded DNA. Which strand will Telomerase extend?
Cytoplasm, nucleus
In prokaryotes, transcription occurs in the ____, whereas in Eukaryotes, it takes place in the _______
False
True or false: Promoters regulate transcription of one bacterial and archeal gene each
Eukaryotic
Do eukaryotic or prokaryotic cells contain introns and exons?
5’ cap addition, intron splicing, and poly-A tail addition
What three additional processes are in transcription in eukaryotes?
RNA polymerase I
Transcribes ribosomal RNA genes. Simpler but fundamental at a cellular level
RNA polymerase II
The most important and complex RNA polymerase, as it transcribes all protein-coding genes into messenger RNA