M2.4: DNA Translation
1/11
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
Types of Ribosomes
PROKARYOTES: Monosome 70s
large subunit: 50s; small subunit: 30s
EUKARYOTES
large subunit: 60s; small subunit: 40s
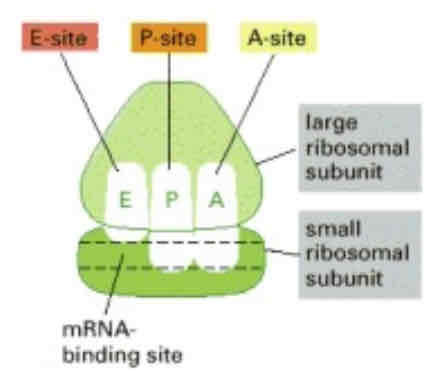
Ribosome: important sites
mRNA binding site
found in small subunit of ribosome
Aminoacyl or Acceptor (A) site: binding site of tRNA + amino acids
Peptidoglycan or Peptide (P) site: tRNA + growing polypeptide
Exit site (E): tRNAs that no longer have a bound amino acid exit from the ribosome
Catalytic site: covalent bond (peptide bond) between 2 amino acids
bond forms between C and N of the 2 aa
Peptide bond synthesis
carbonyl end of the 1st amino acid and the amino end of the 2nd amino acid are connected
Translation stages
initiation
elongation
termination
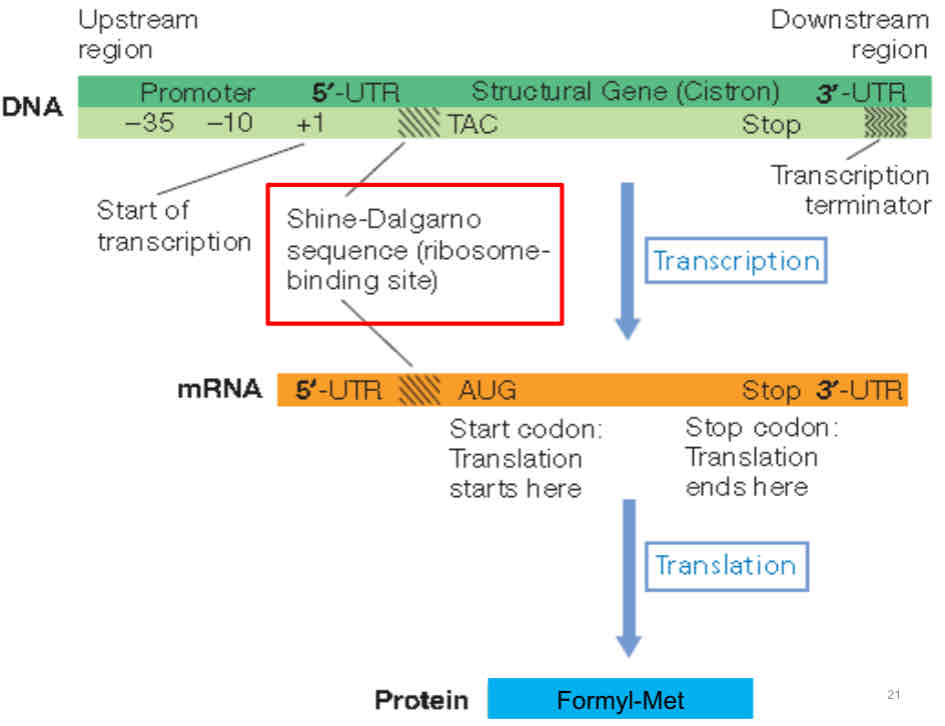
Shine dalgarno sequence
ribosomal binding site that serves as an initiating signal for protein synthesis
enables initiation of protein synthesis by aligning the ribosome with the start codon
indicates where protein synthesis starts, thus ensuring that genes are read correctly
found between untranslated region and sequence coding for the 1st aa (which explains why proteins are generally shorter than mRNA: since shine dalgarno sequence is not translated)
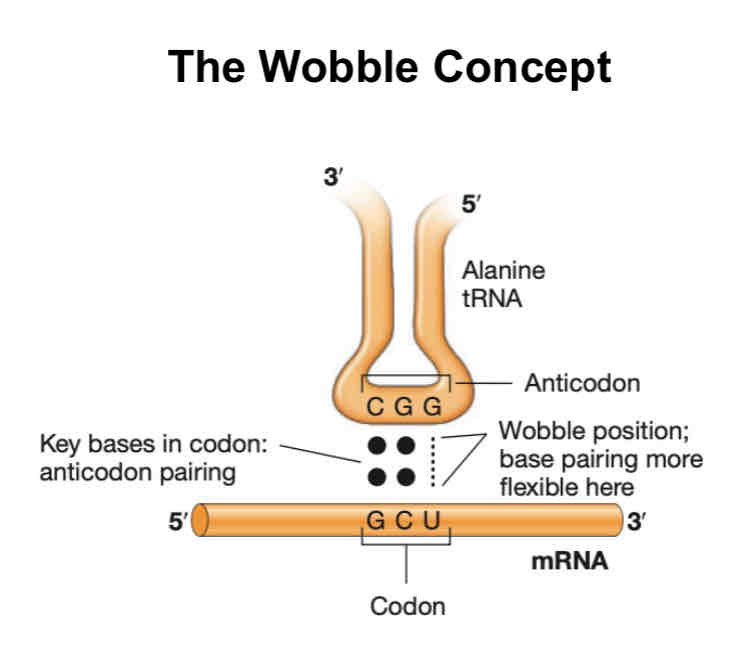
Wobble concept
codons don’t follow specific base pairing
no need for 1:1 correspondence between codons and tRNA (1 tRNA can recognize multiple codons which code for the same sequence)
Initiation (EUKARYOTES)
ribosome, mRNA, and initiator tRNA contains the 1st aa in the protein (usually Met) come tgt to form the initiation complex
tRNA containing Met attaches to the small ribosomal subunit. tgt, they bind to the 5’ end of the mRNA by recognizing the 5’ GTP cap (added during processing in the nucleus)
afterwards, tRNA + small ribosomal subunit go down the mRNA in the 3’ direction, stopping when they reach the start codon (AUG)
Initiation (PROKARYOTES)
small ribosomal subunit attaches to the shine-dalgarno sequences (which is just before the start codons)
Elongation
Met-carrying tRNA starts in the P site (middle slot of the ribosome). next to it, a fresh codon is exposed in the A site
A site will be the “landing site” for the next tRNA, one whose anticodon is a complementary match for the exposed codon
once the complementary tRNA has landed in the A site, a peptide bonds will form between the amino acids and connect them to each other (Met will be transferred frm the 1st tRNA to the aa of the 2nd tRNA in the A site)
Met forms the N-TERMINUS of the polypeptide (end of the polypeptide with an exposed amino group)
other aa is the C-TERMINUS (end of the polypeptide with an exposed carboxyl group); new aa are progressively added to the C-terminus
once a peptide bond is formed, the mRNA is pulled onward through the ribosome by 1 codon; this shift allows the 1st empty tRNA to drift out via the E site and also exposes a new codon in the A site, thus allowing the cycle to repeat
Termination
stop codons are recognized by release factors (proteins which fit into the P site)
release factors disrupt the enzyme that normally forms peptide bonds (they make it add a H2O molecule to the last aa of the chain, which separates the chain from the tRNA and releases the newly-synthesized protein)
after the small and large ribosomal subunits separate from the mRNA and from each other, each element can take part in another round of translation
Eukaryotic translation
translation is separated spatially and temporally from transcription (since sites for transcription and translation are separated by organelles)
larger ribosomes
longer rRNA (expansion segments needed for ribosome assembly and translation regulation and specificity)
expansion segments are not coded by DNA/not part of the gene
no shine-dalgarno sequence
instead use cap-dependent translation
initiation starts with the small subunit associating with m^7 G cap (CAP-DEPENDENT TRANSLATION)
eukaryotic initiation factors (eIFs), eukaryotic elongation factors (eFFs), eukaryotic release factors (eRFs)
tRNAiMet: unformylated Met (which is why first amino acid is always fMet)
presence of Kozak sequence (A/GNNAUGG): increased efficiency of translation initiation
has purine base (adenine/guanine); then any other base
enhances efficiency of translation initiation
eukaryotic mRNAs (10 hrs) are longer lived than bacterial mRNAs (5 mins)
can live longer without being degraded)
Poly-A binding proteins are required to form the initiation complex
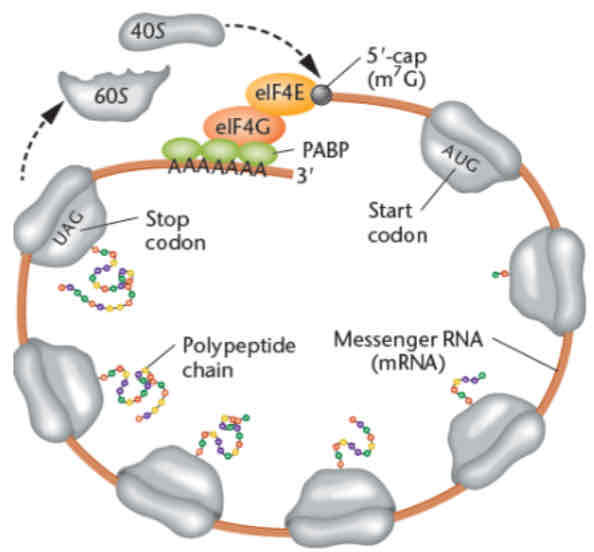
closed-loop translation
avoids wasting energy in translating partially degraded mRNA
efficient ribosome recycling