Evolution - Exam 2 - All Key Terms (C4-C5) + Some Questions on Concepts
1/80
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
81 Terms
Hardy-Weinberg equation
p2 + 2pq + q2 = 1
p+q=1
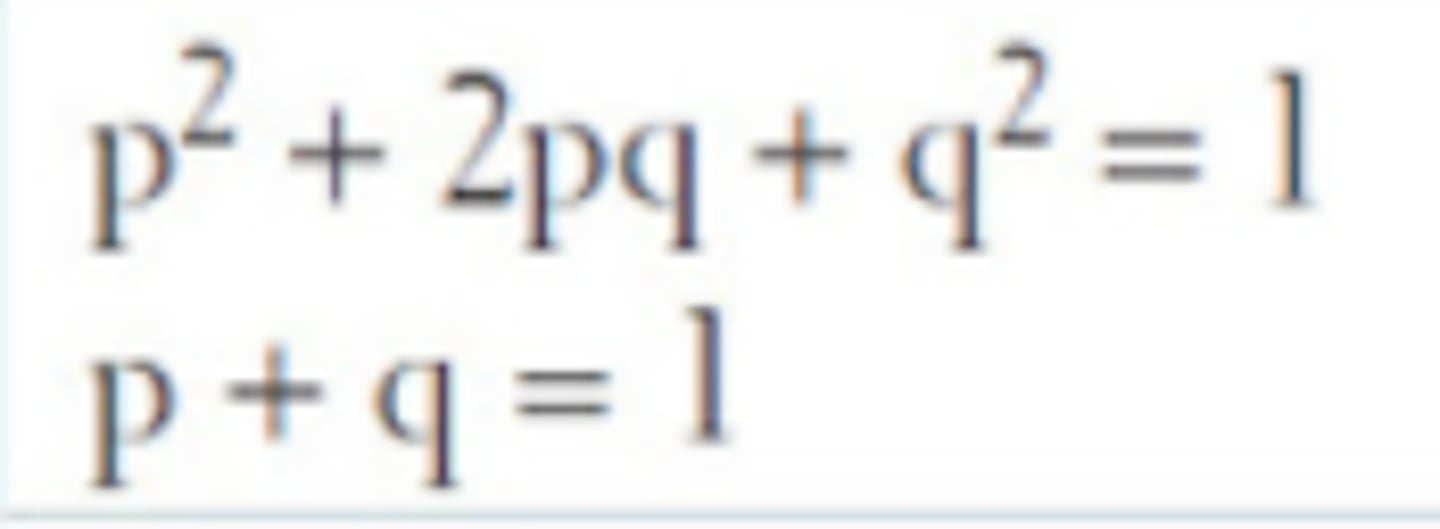
fitness components equation
W = (probability individual survives to maturity) x (expected number of offspring if individual survives)
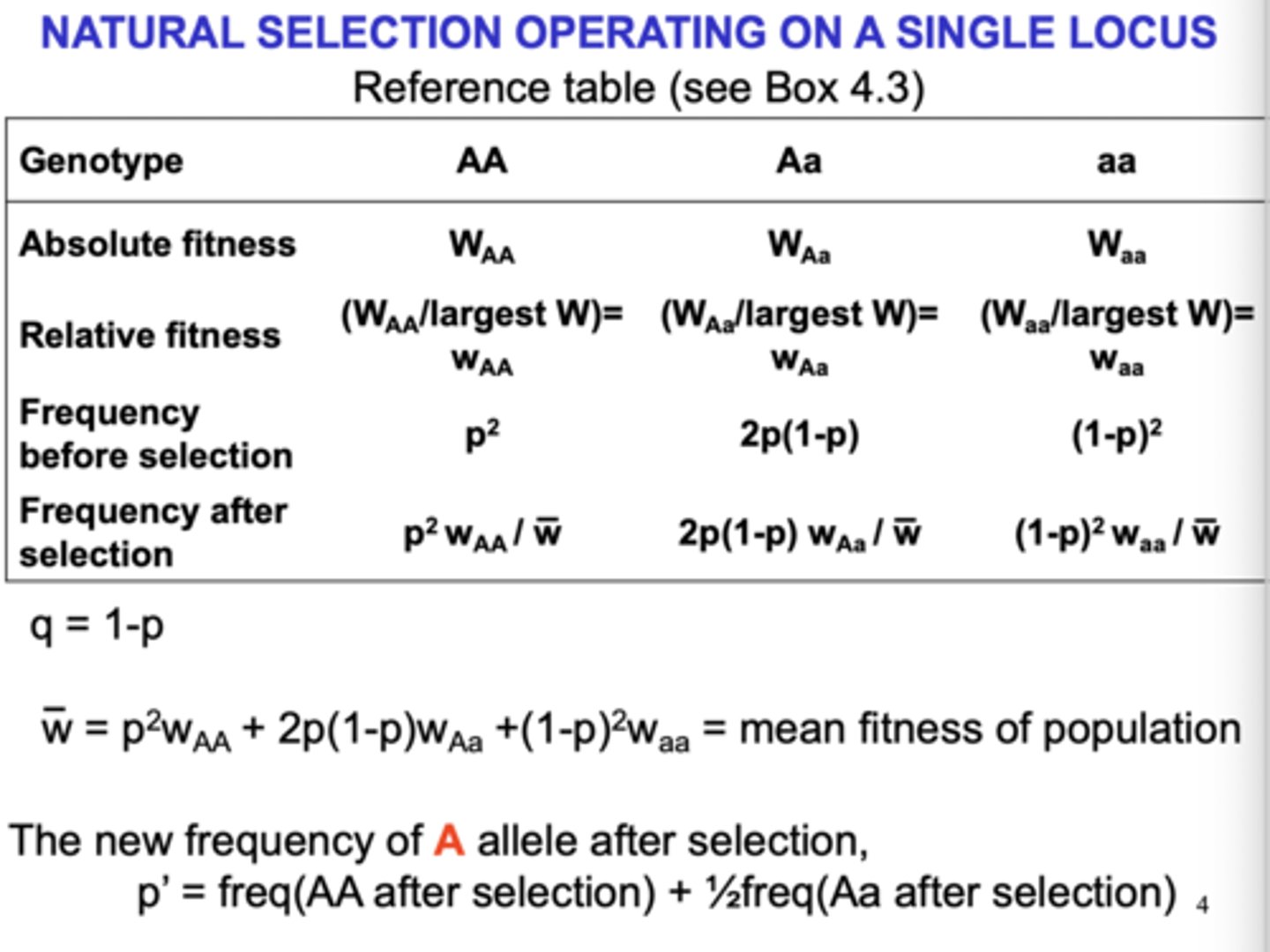
selection on a single locus
delta P = sp(1-p)/w = sp(1-p)
(Notes on what each means:)
(p = frequency of A2)
(delta p = change in the rate of frequency of the allele)
(w = mean fitness)
(s = selection coefficient of A2)
(1-p = frequency of A1)
mutation selection balance
p = m/s
(when mutation adding harmful alleles is equal to selection removing them, allele frequency stops changing)
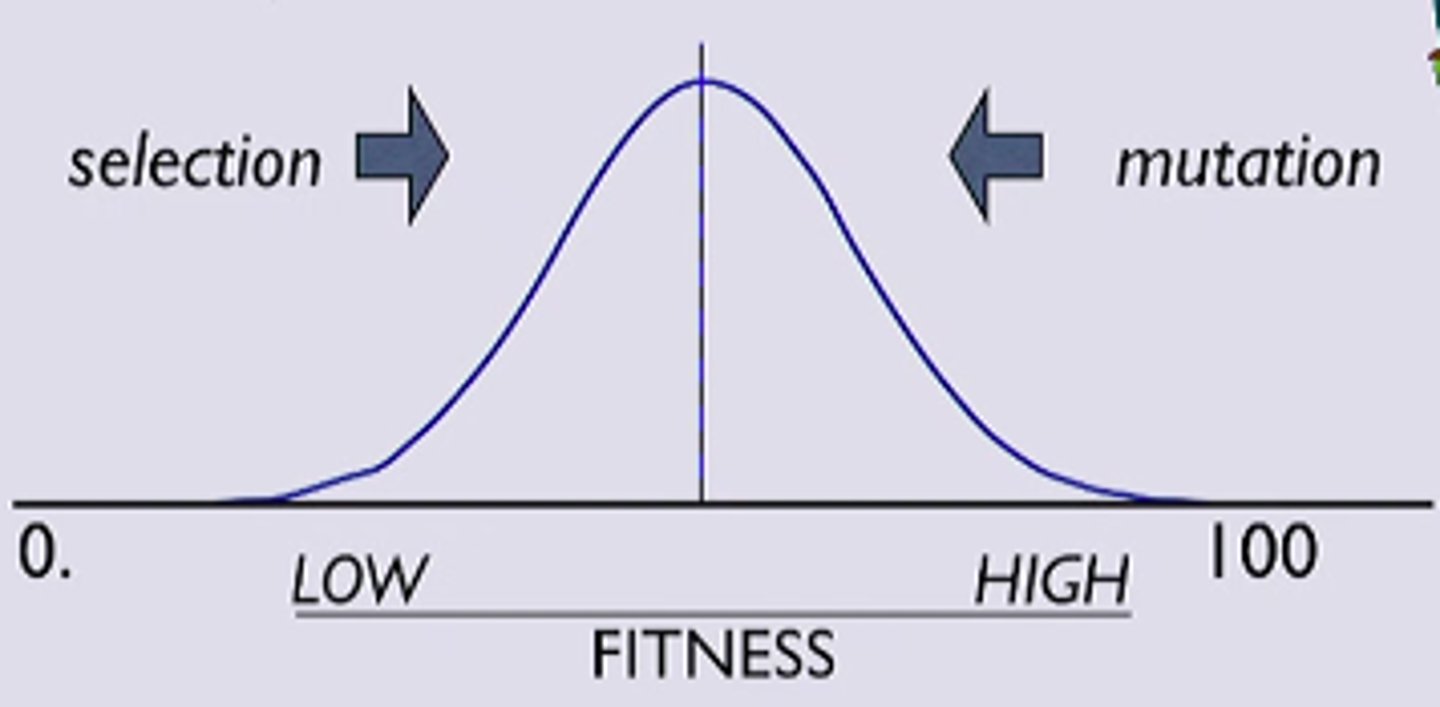
pr (fixation)
pr (fixation) = 2s
(calculates the time for an allele to reach frequency of 1)
allele
different forms of the same gene
allele frequency
the chance of randomly picking an allele from a population
beneficial mutation
enhances the survival or reproductive success of an organism

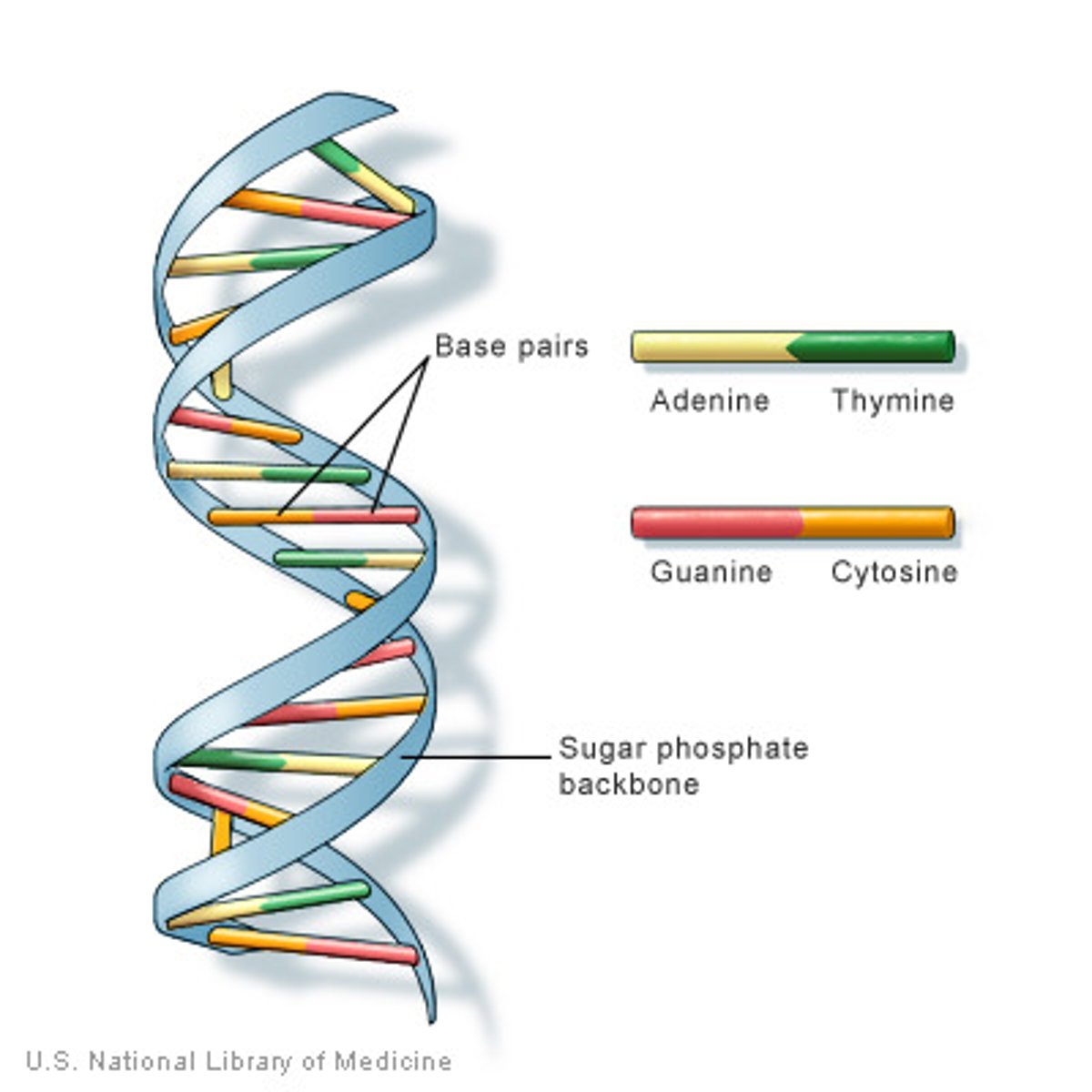
base pair (bp)
a pair of either A-T or C-G (Adenine-Thymine or Cytosine-Guanine)
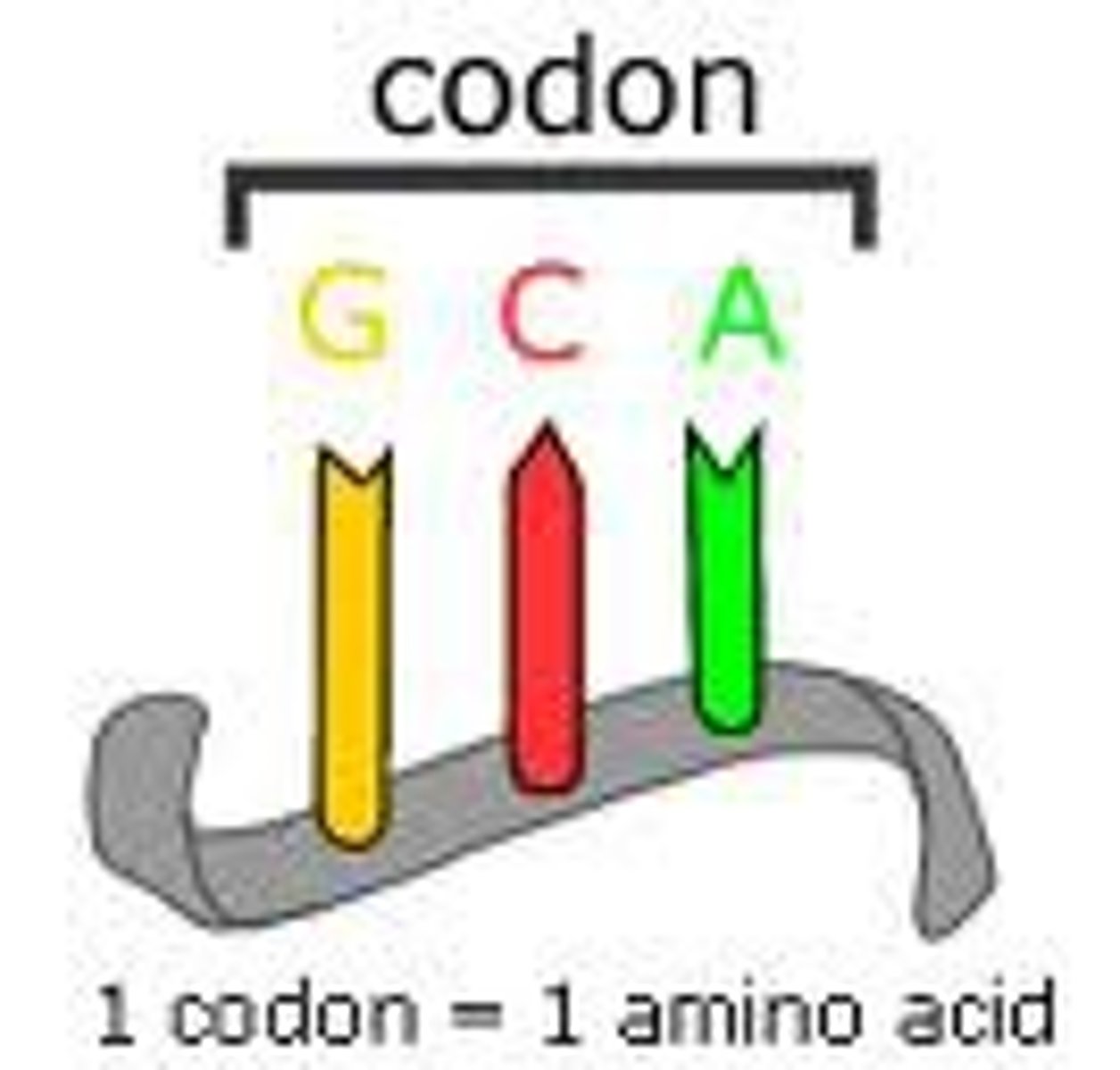
codon
a set of three bases (ex: ATG) that codes for an amino acid or stops transcription
cultural inheritance
traits transmitted by behavior and learning

deleterious mutation
genetic changes that are harmful to an organism.

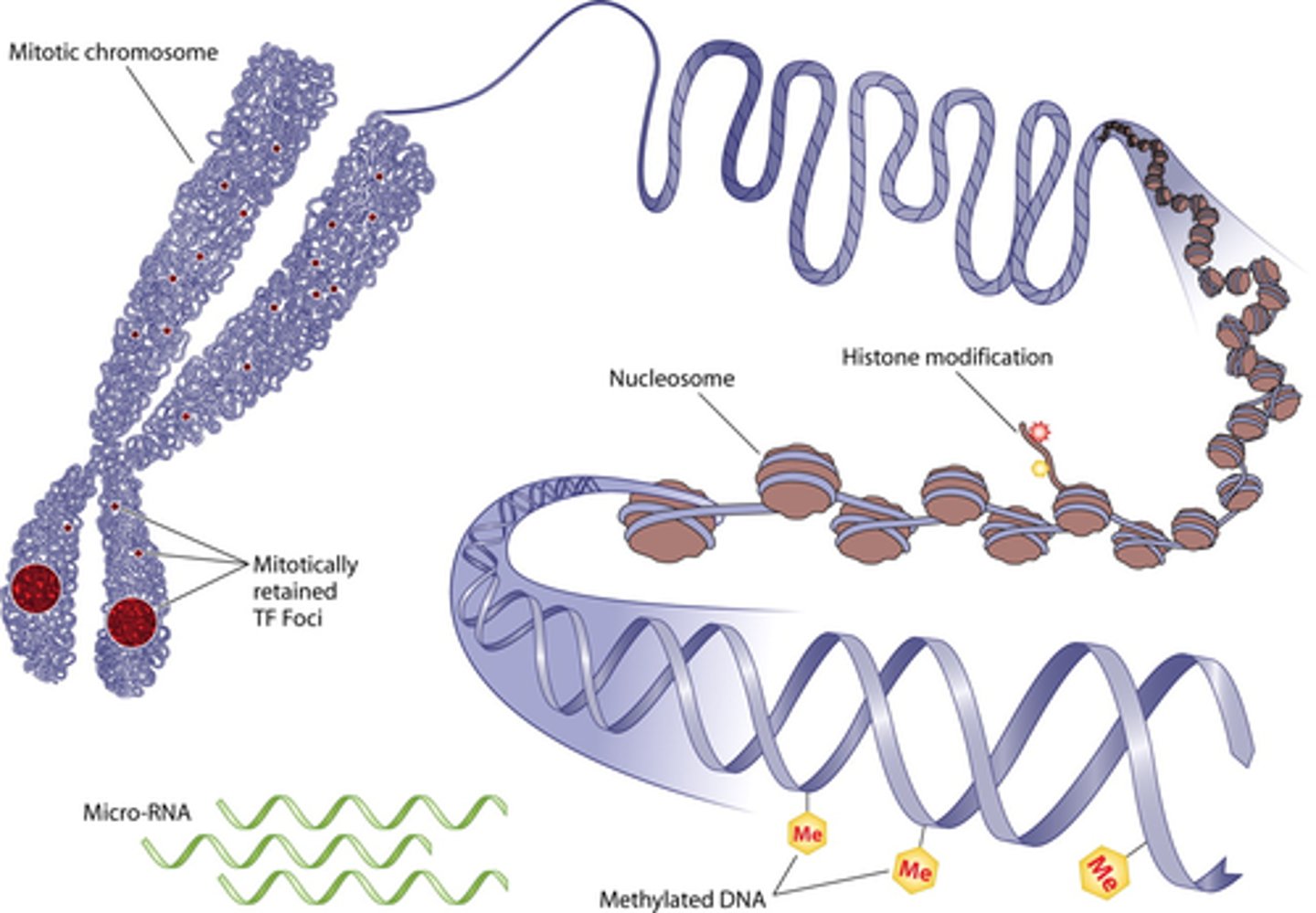
epigenetic inheritance
when offspring inherit phenotypic traits from their parents by other means (not mutations in DNA)
epistasis
when the effect of one gene on a trait depends on the presence of another gene

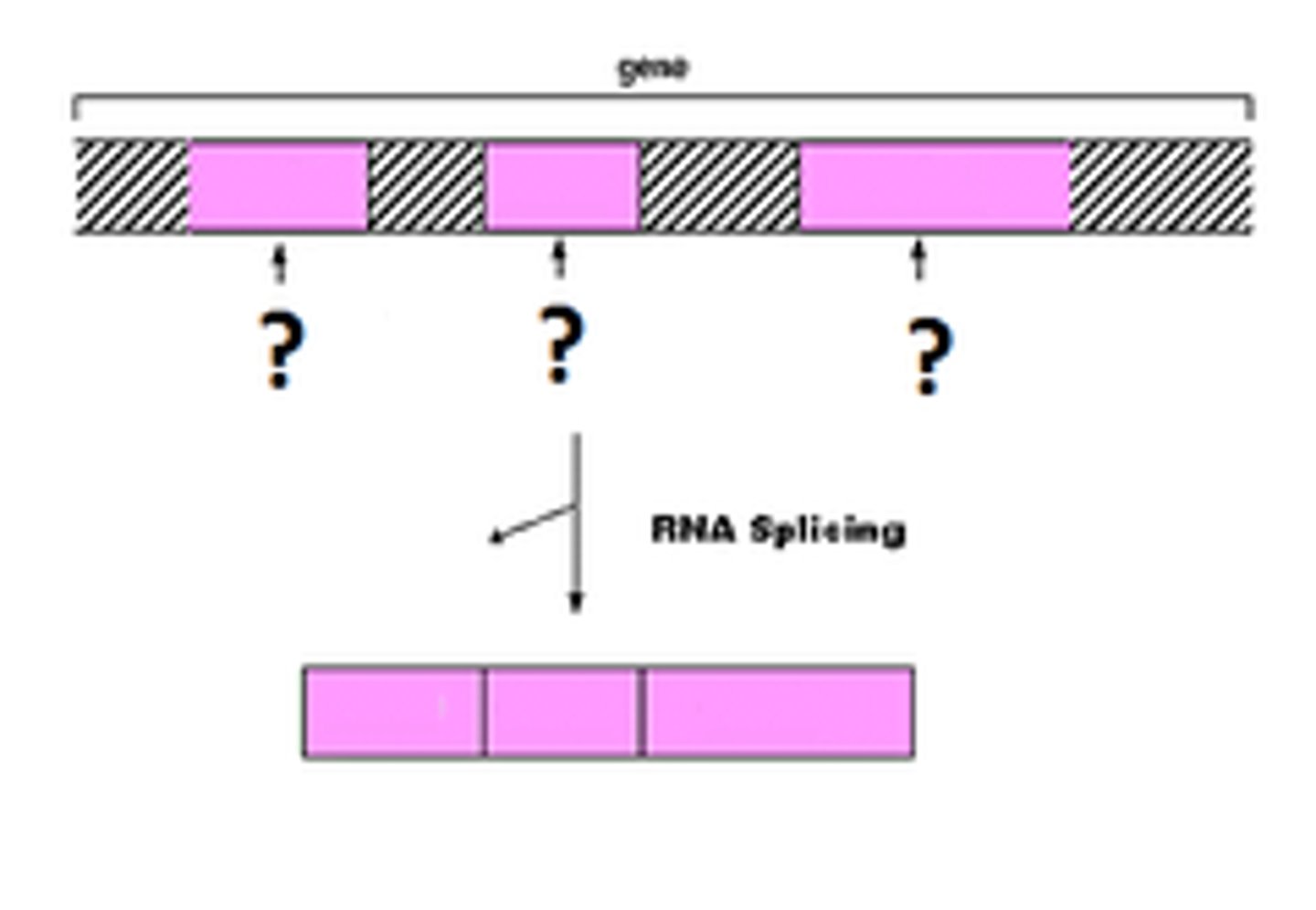
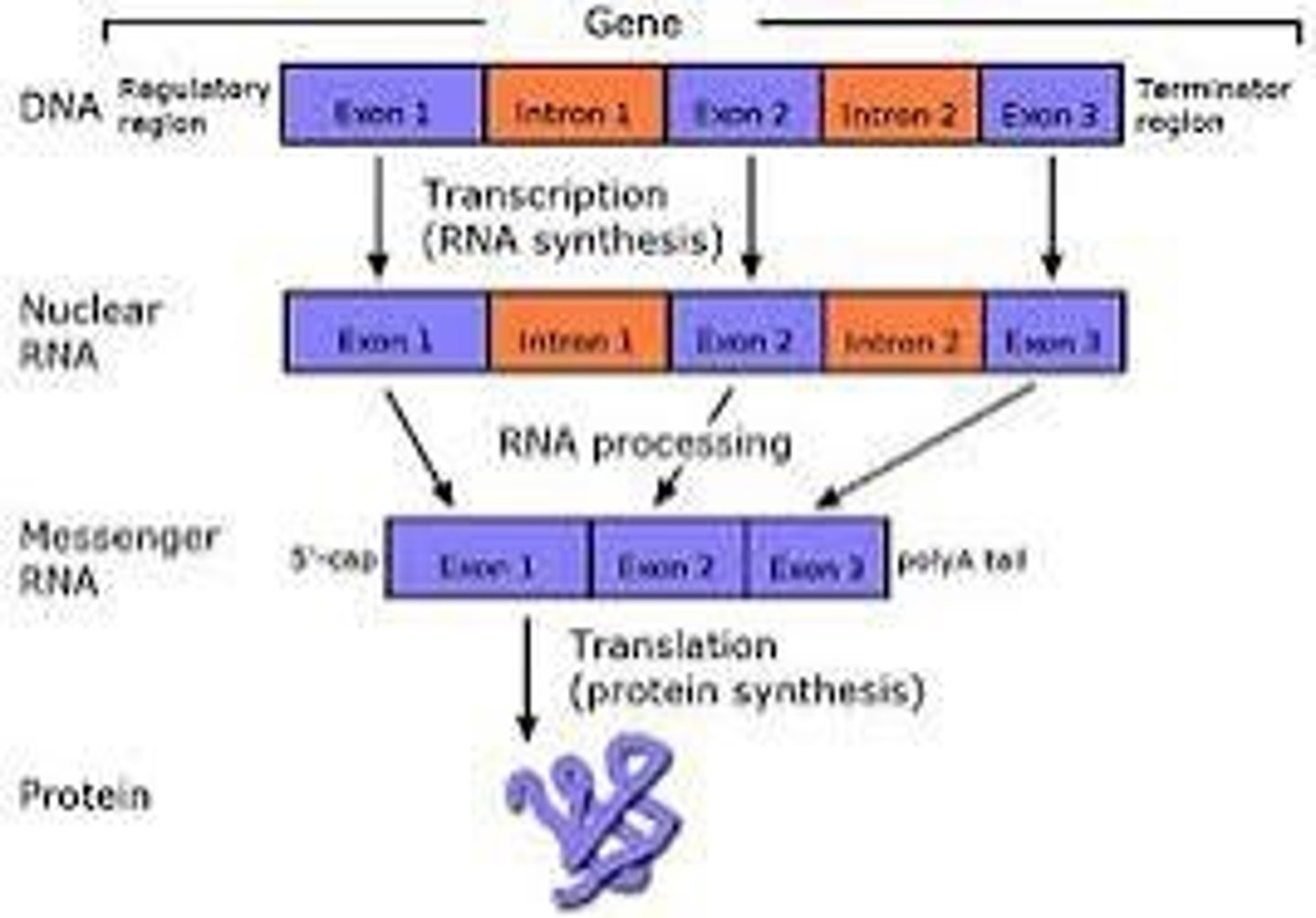
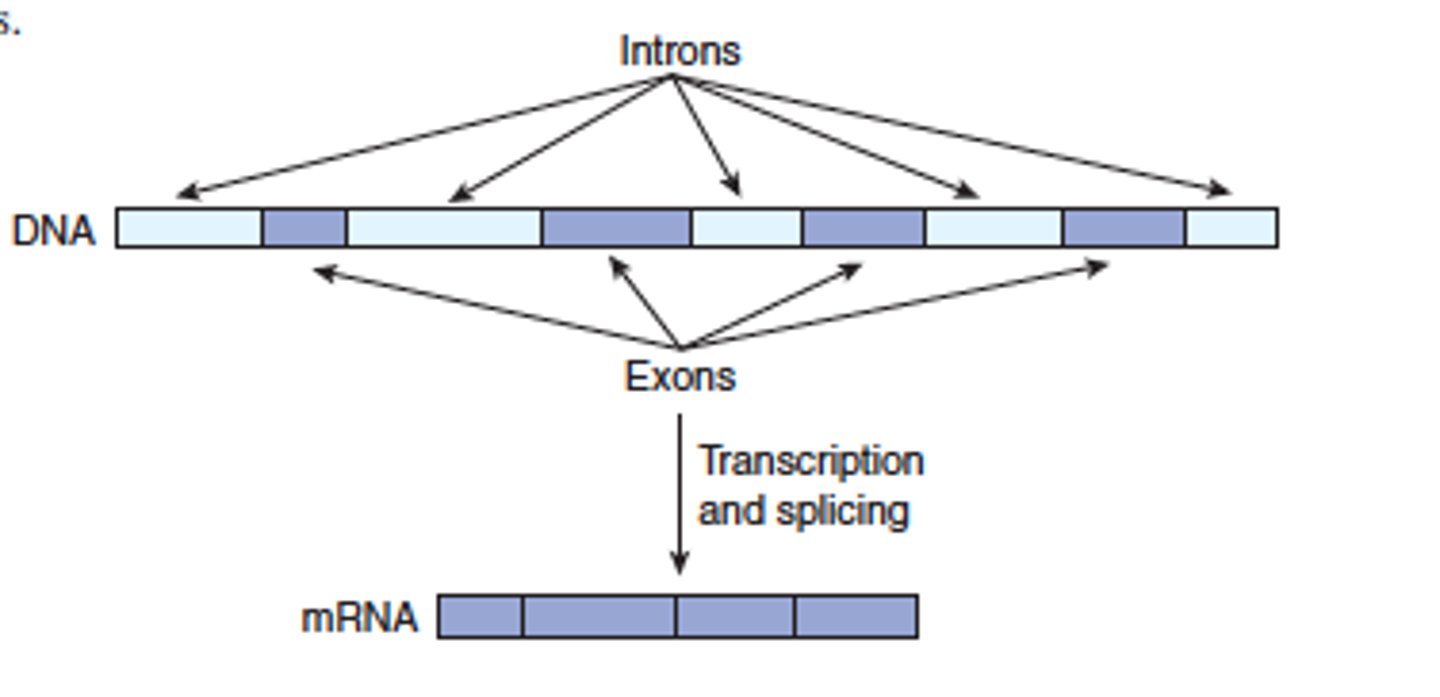
exons
segments (left over after removing the introns) joined together to make the final RNA
fitness
the number of offspring an individual leaves to the next generation.
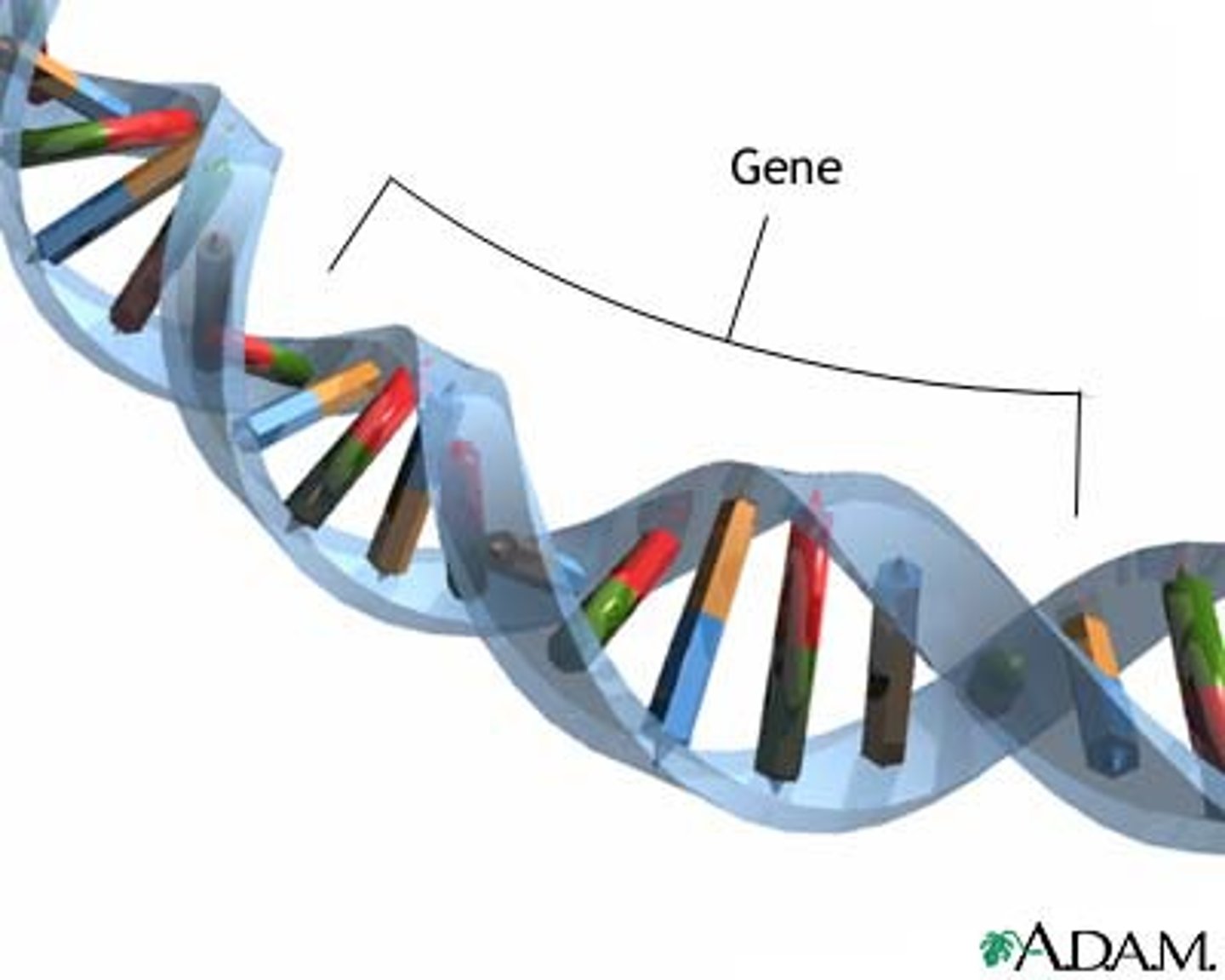
gene
segments of chromosomes that perform a function (like coding for proteins)
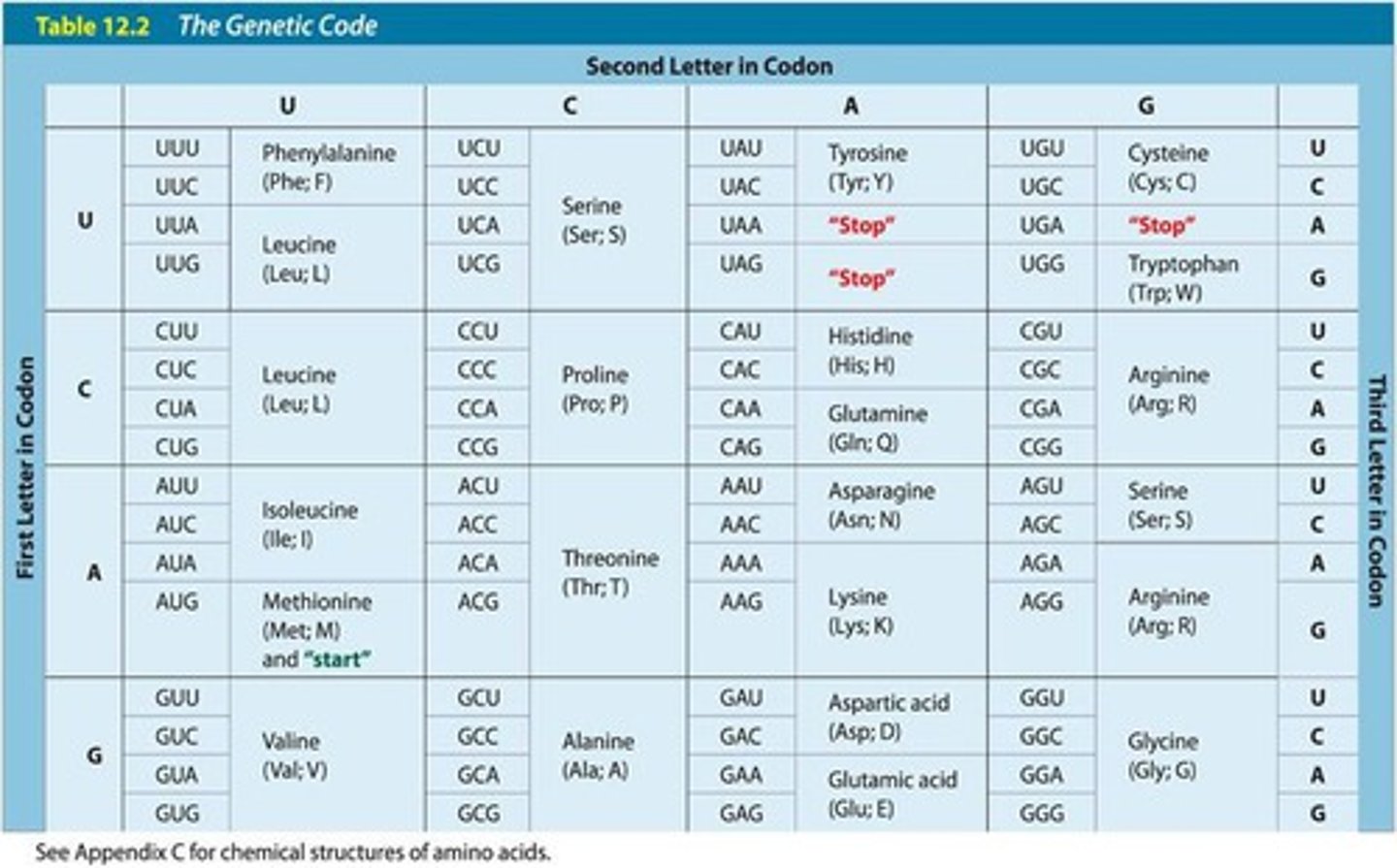
genetic code
map showing how all possible codons correspond to an amino acid
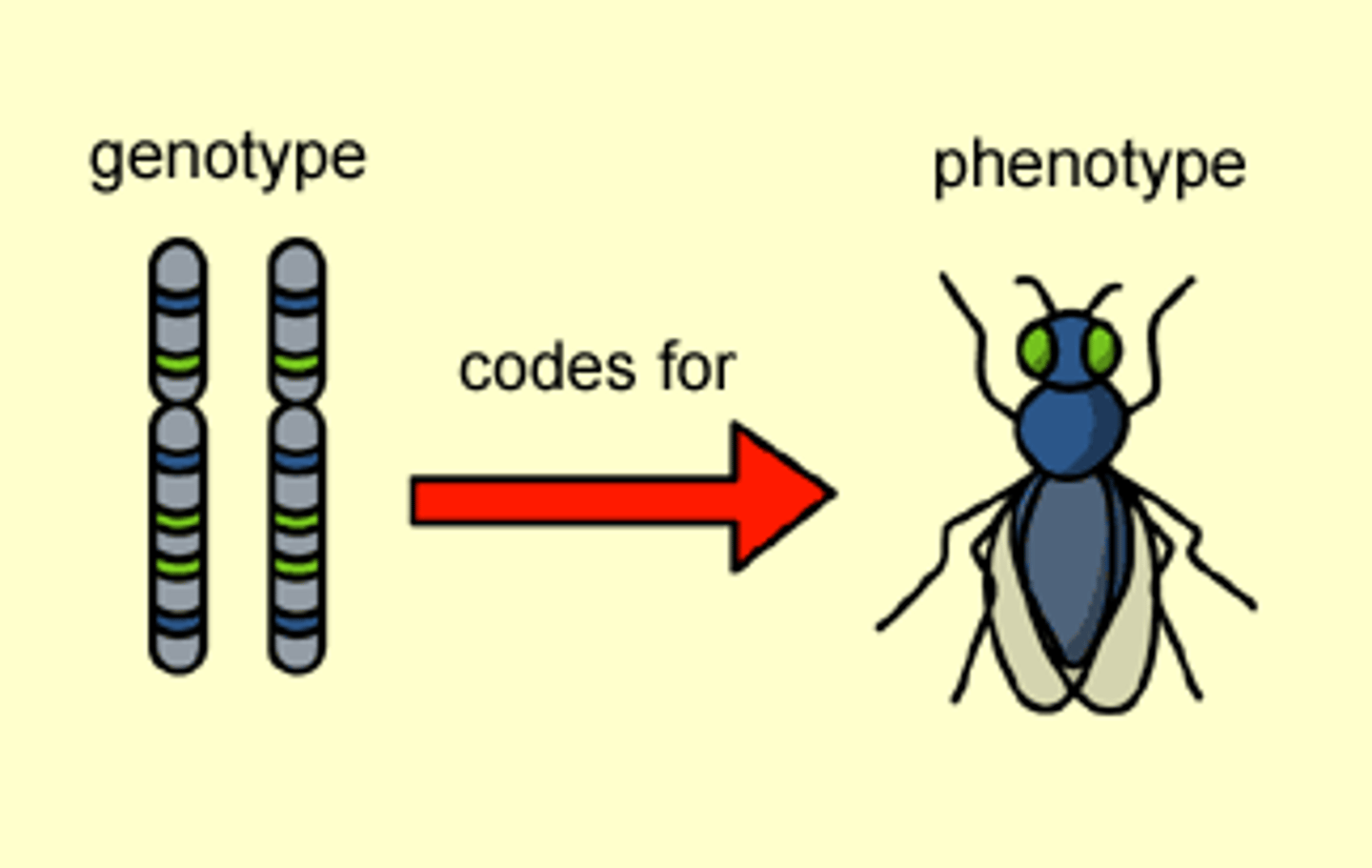
genotype
the combination of genes an individual has for a specific trait
germ line
cells that give rise to gametes (eggs and sperm)
Hardy-Weinberg equilibrium
when a population's genetic makeup remains stable over generations with no evolution occurring (requires no mutations, migrations, selection, or genetic drift)
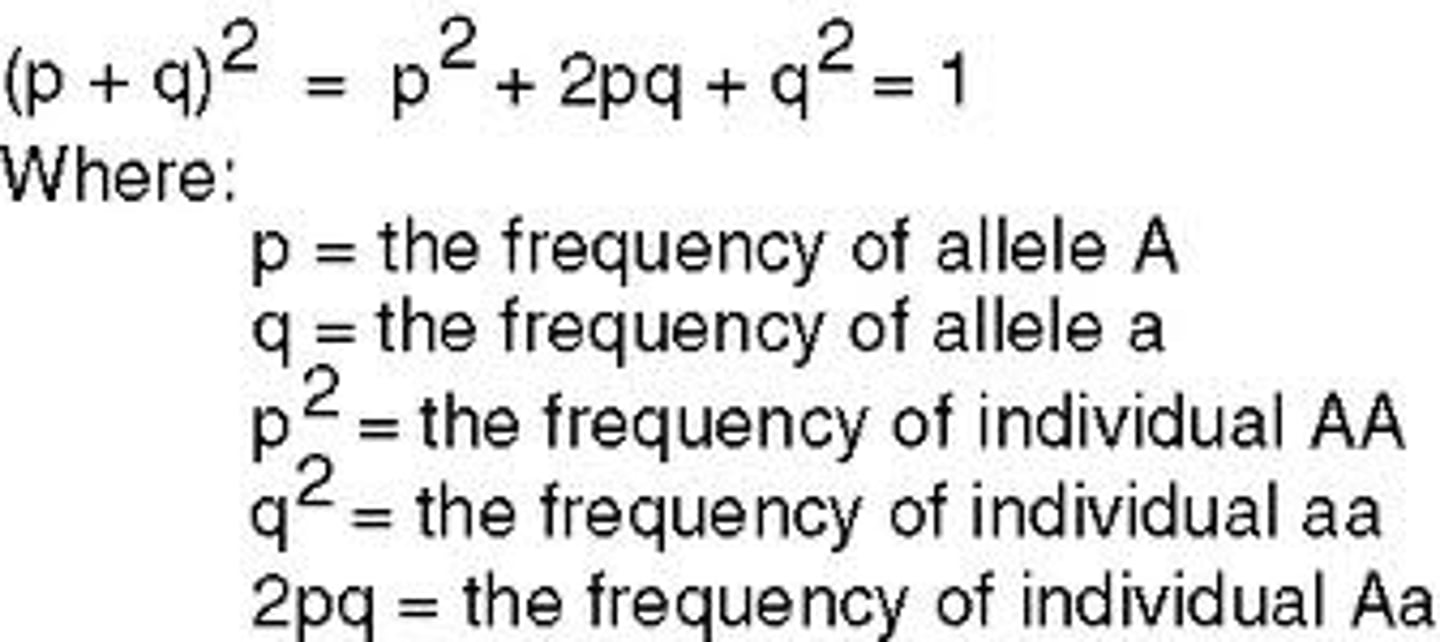
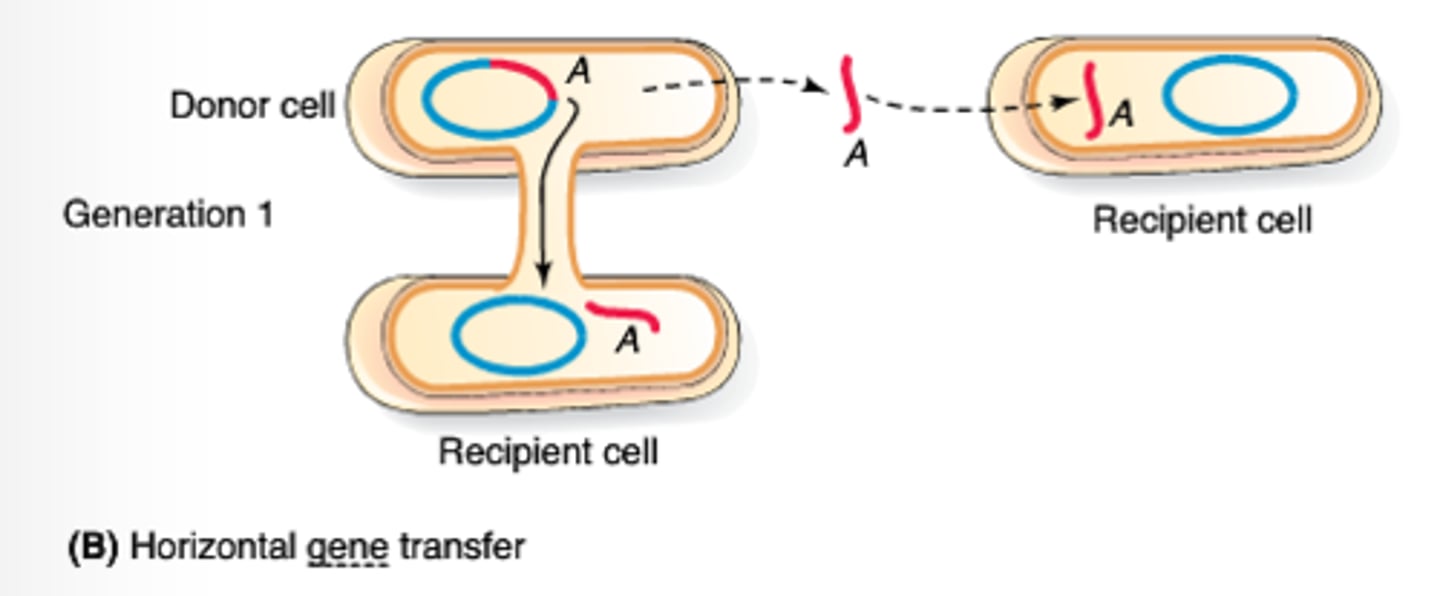
horizontal gene transfer (HGT)
the movement of DNA between different individuals without sexual reproduction
intron
segments removed from the pre-mRNA (does not code for amino acid)
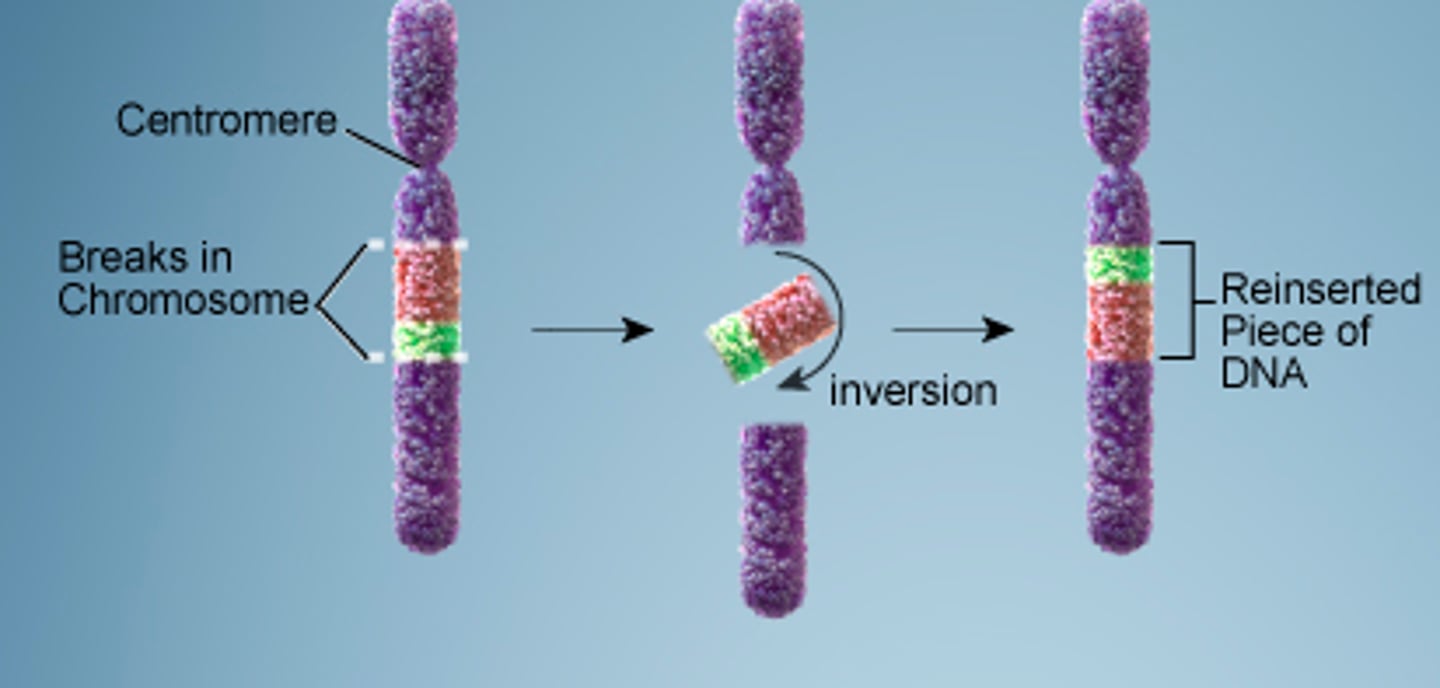
inversion
when a chromosome breaks in two places and the middle segment is reattached in the reverse orientation
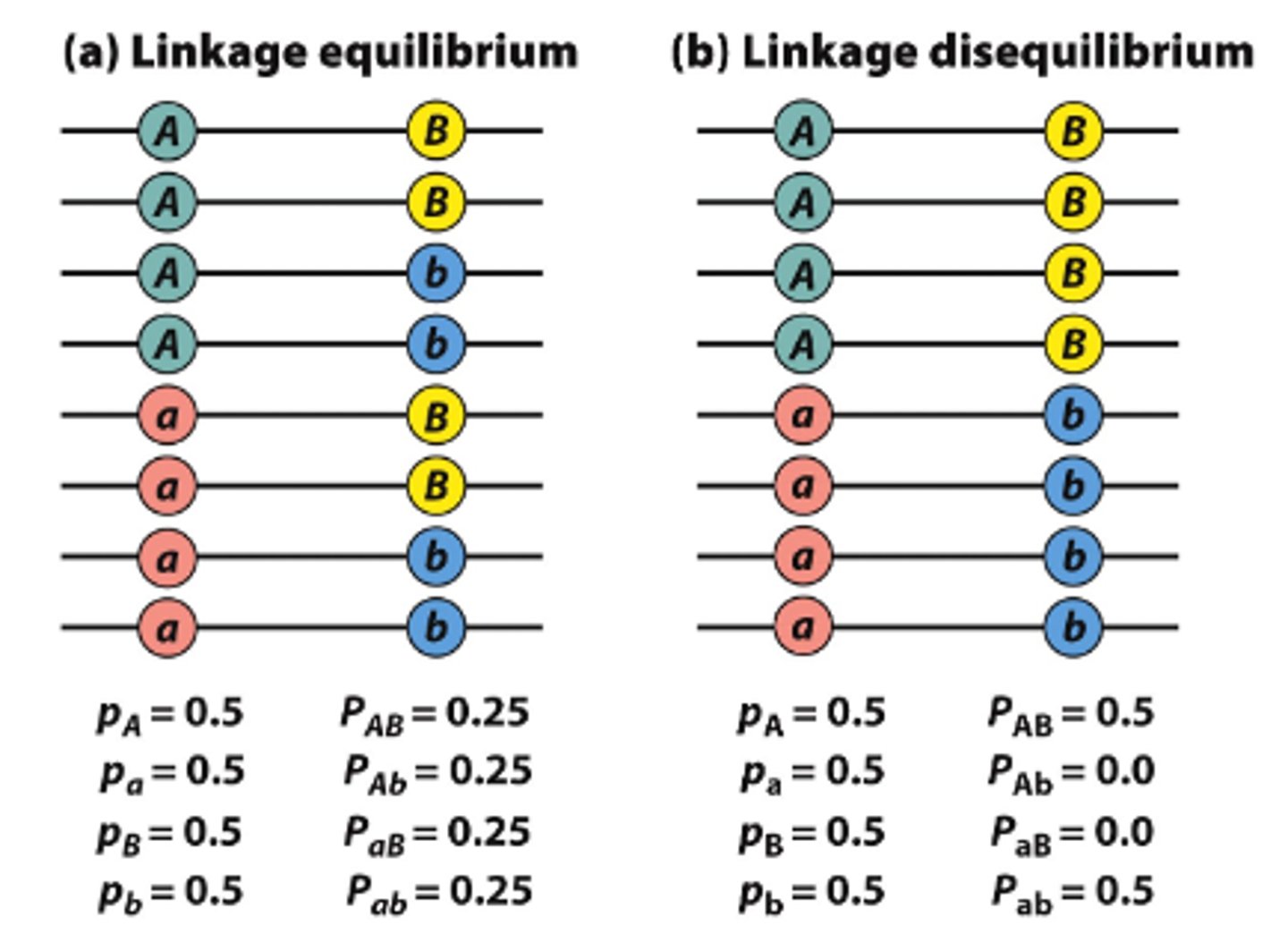
linkage disequilibrium
when two alleles are found together more often than expected by chance
Can linkage disequilibrium still occur even if loci are not physically linked?
Yes, still occurs.
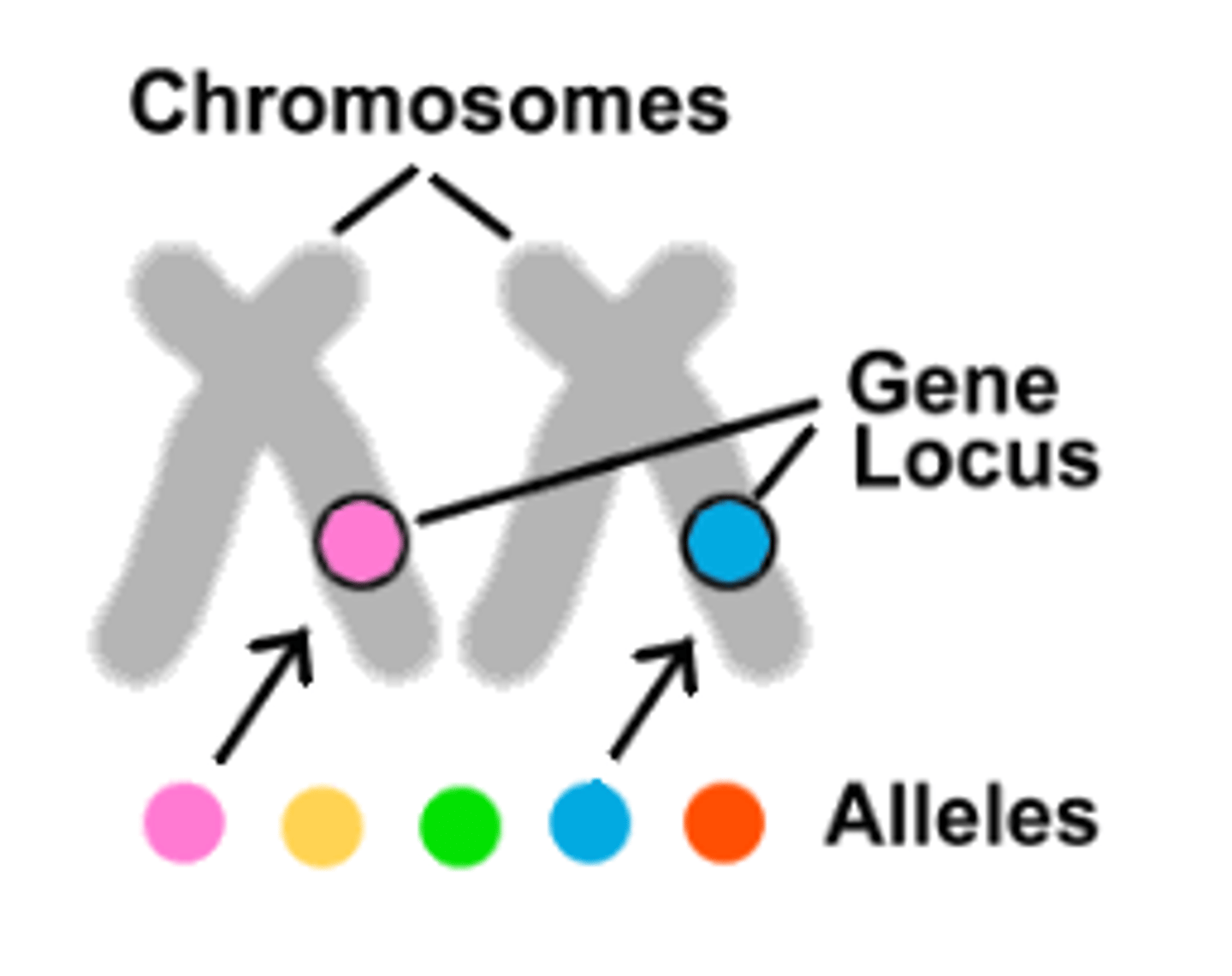
locus
site on a chromosome where a gene is located
maternal effect
when the genotype or phenotype of the mother directly influences the phenotype of her offspring

mutation
changes in the DNA or RNA sequence caused by errors during replication
mutation rate
how often a genetic change happens in an offspring compared to its parents
noncoding DNA
regions that don't code for proteins (includes introns)
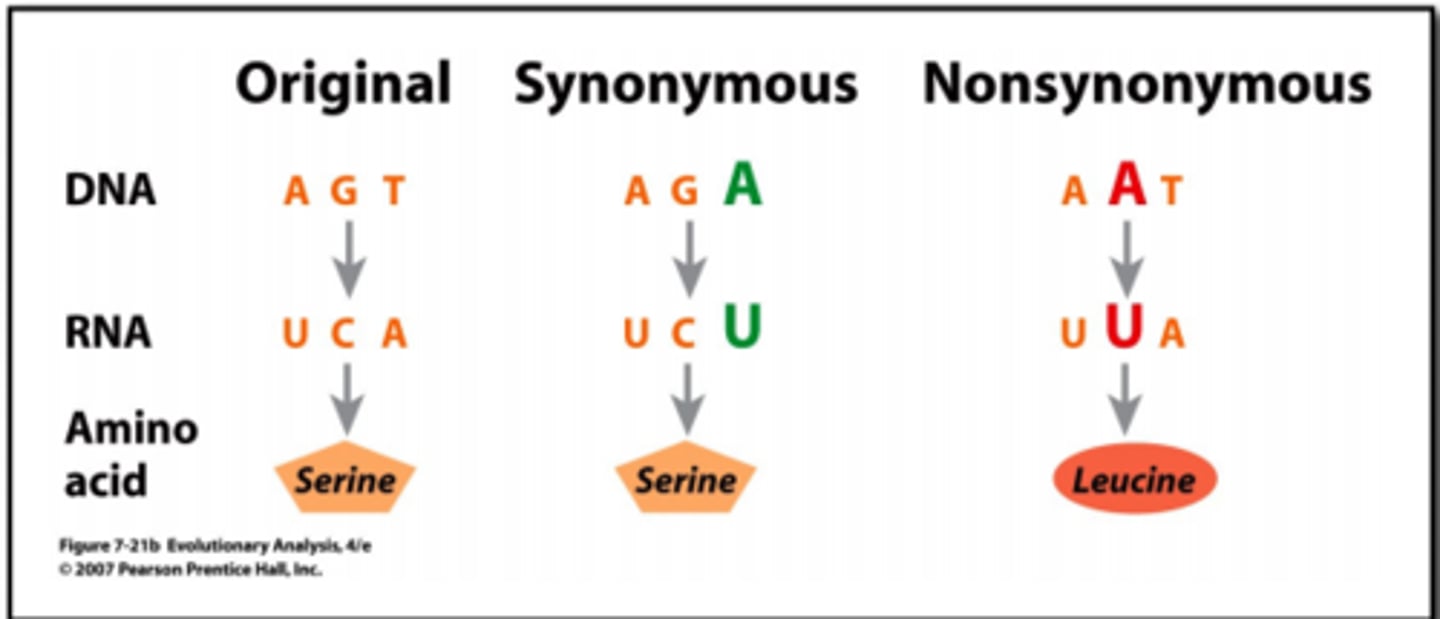
nonsynonymous
a change in a codon that alters the amino acid it codes for
phenotype
observable and visible differences among individuals of a species
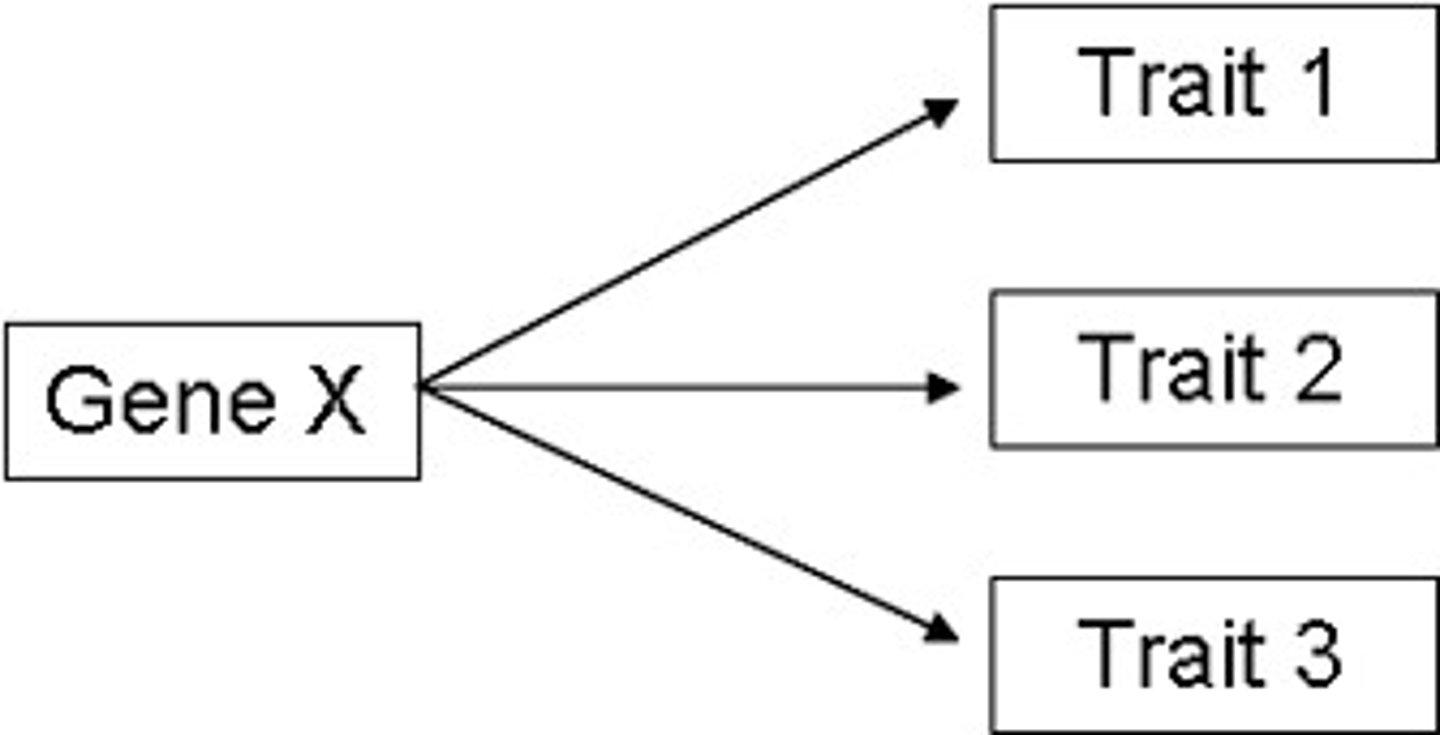
pleiotropy
when a single mutation affects multiple traits (ex: dwarfism)
point mutation
when one single DNA base is swapped for another

polymorphic
a gene or trait existing in two or more forms

recombination
when genes from different parents mix together to create new combinations in offspring
recombination rate
the probability that recombination occurs between two loci (genes)
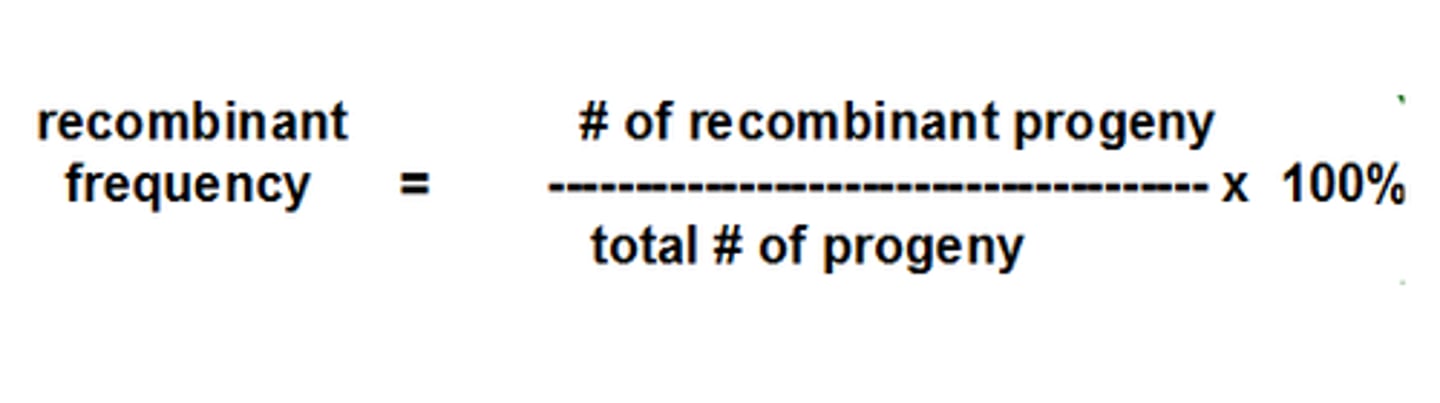
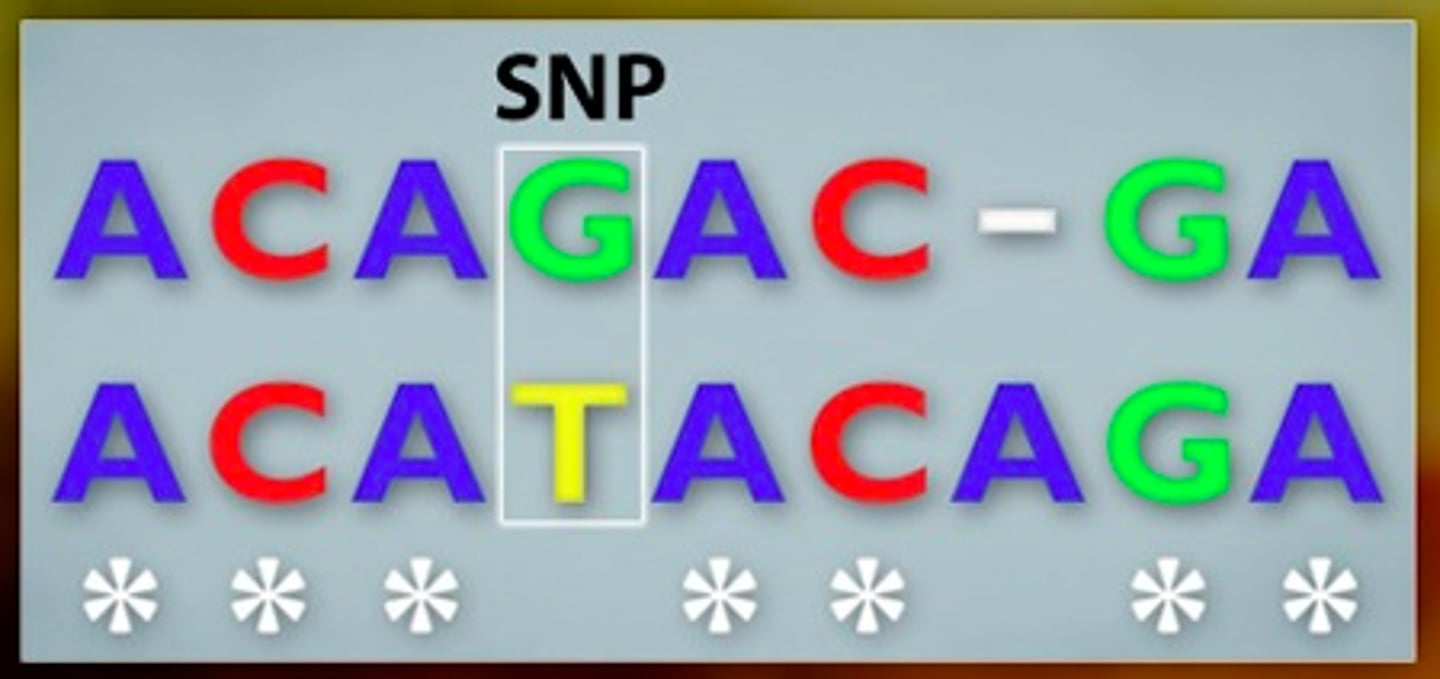
single nucleotide polymorphism (SNP)
when a single DNA base (A, T, C, or G) varies at a specific spot in the genome
soma
all of the body (except the germ line)
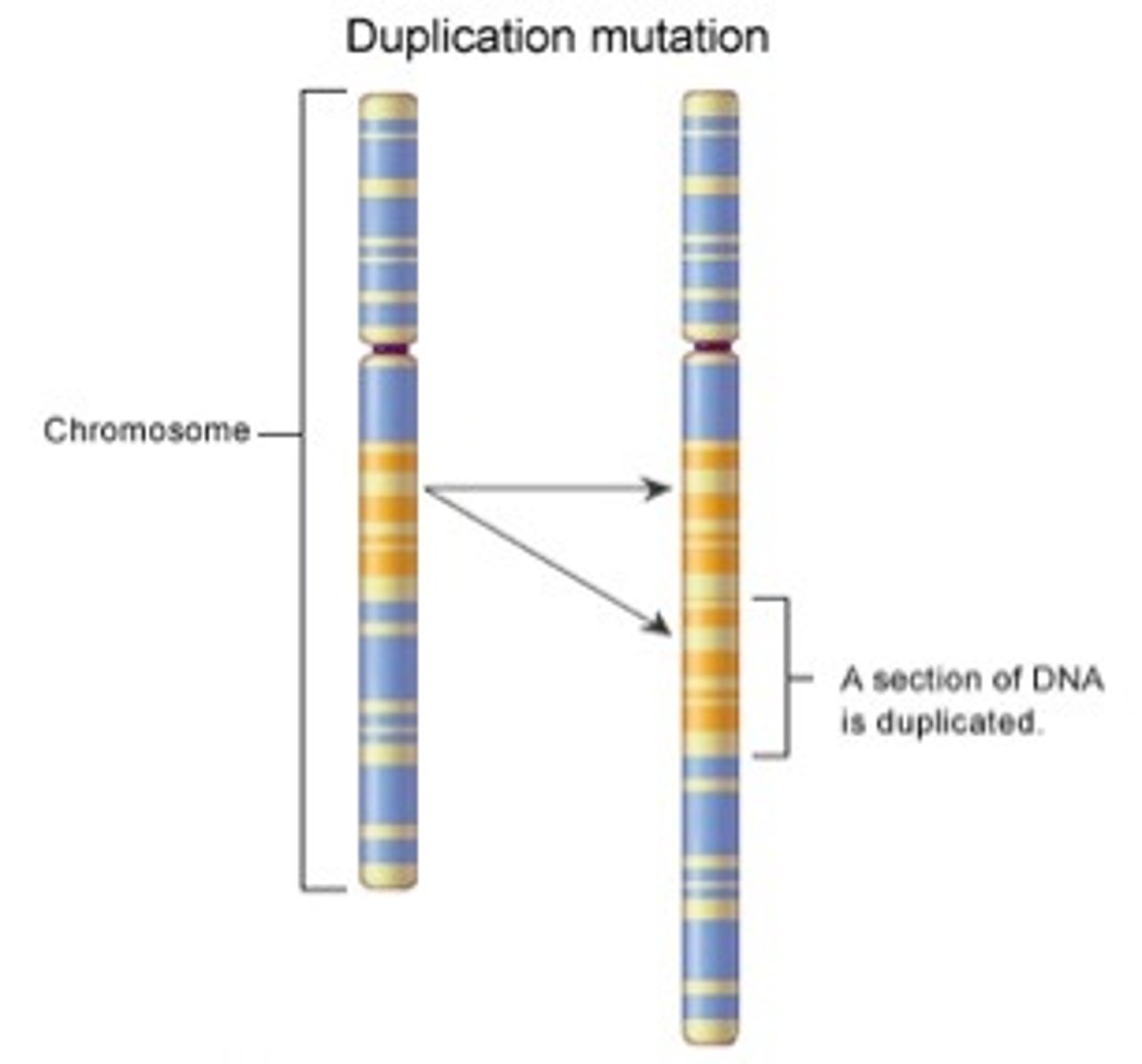
structural mutation
a mutation that changes more than one DNA base in a chromosome
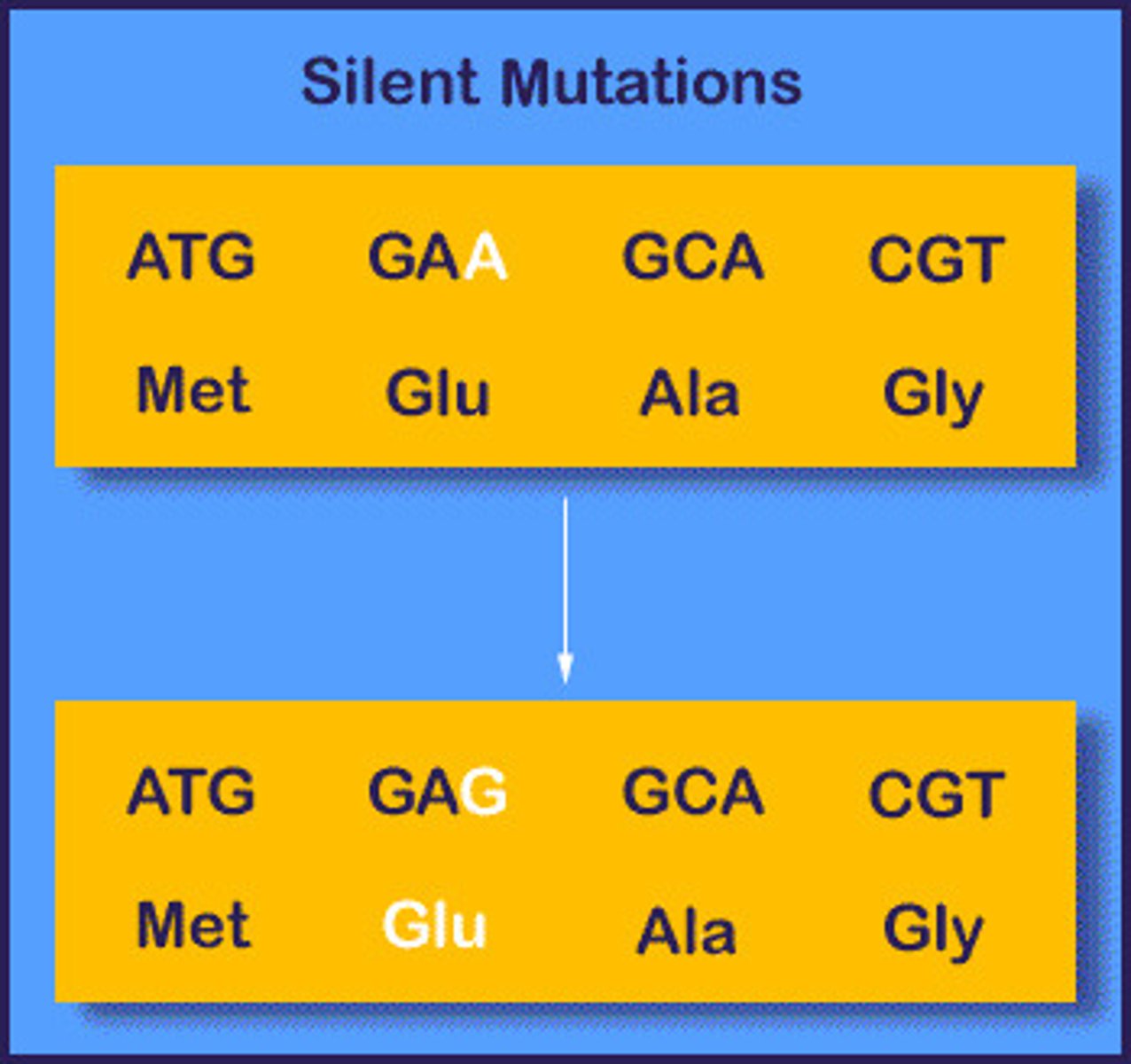
synonymous mutation (silent mutation)
a change in a codon that does not affect the amino acid it codes for
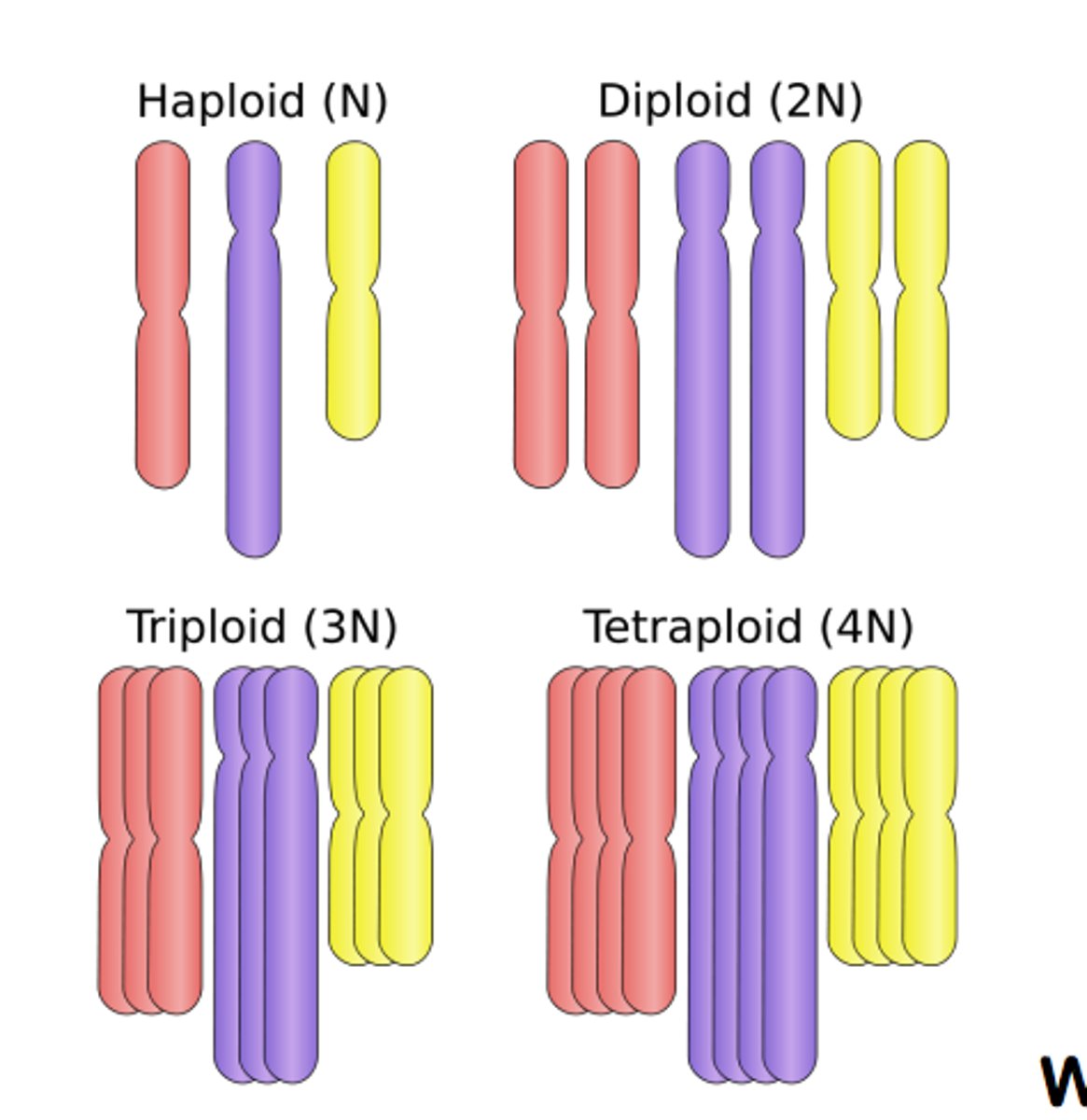
whole genome duplication
when an organism inherits an entire extra set of chromosomes due to errors in meiosis
absolute fitness
number of offspring (zygotes) produced over its lifetime
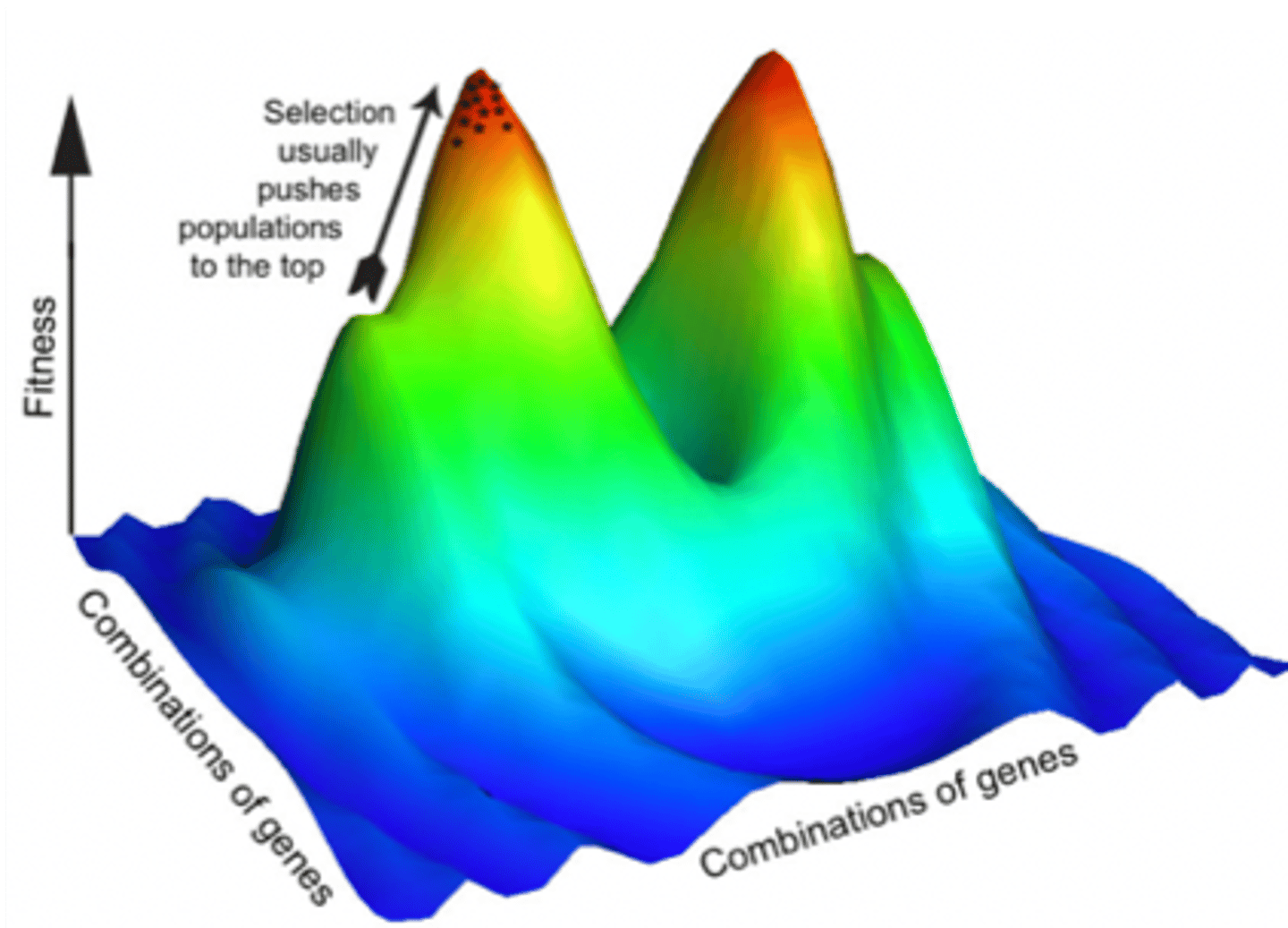
adaptive landscape
when mean fitness is shown as hills and valleys, showing how selection pushes populations "uphill" to higher fitness (x = allele frequency, y = mean fitness)
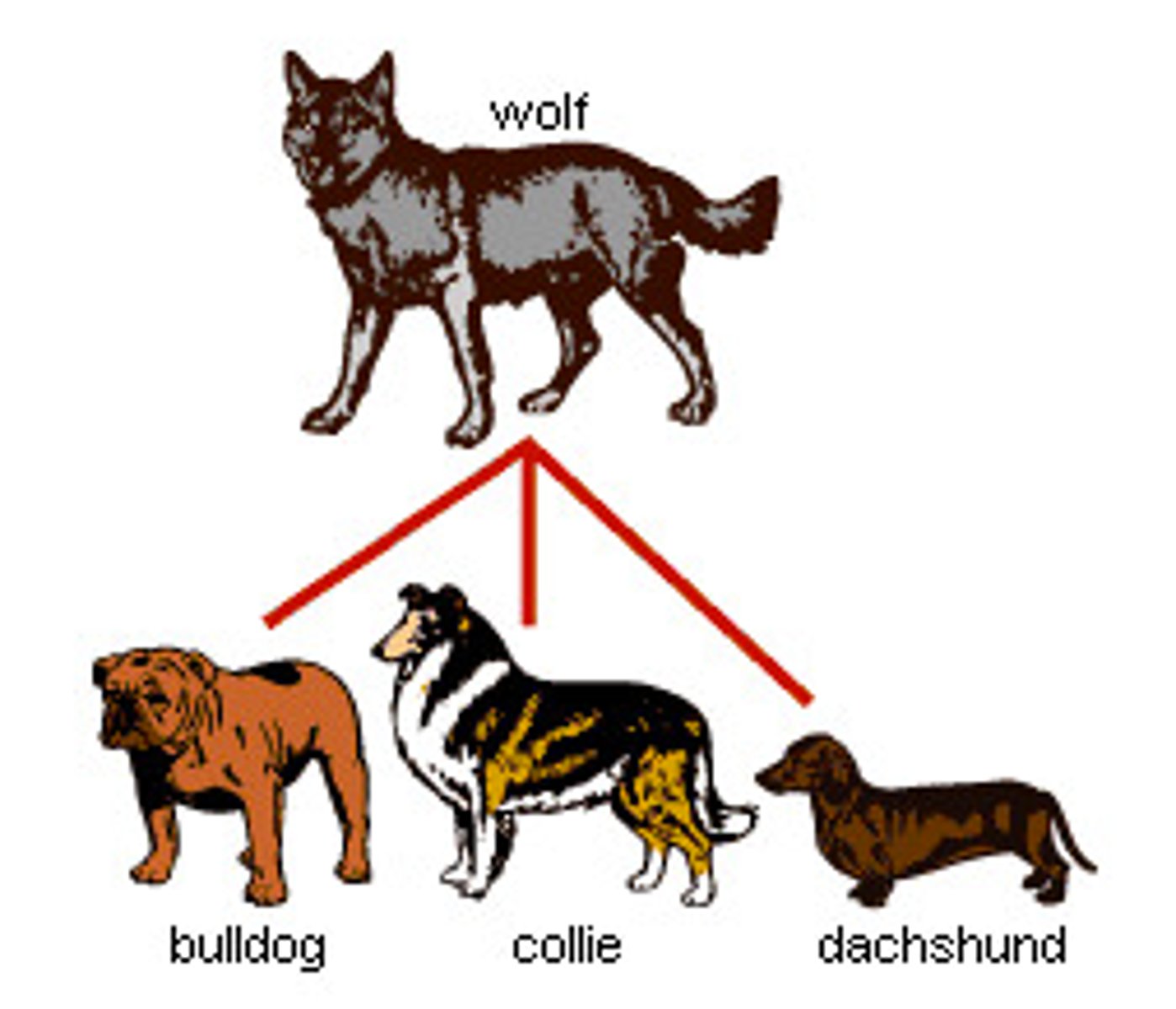
artificial selection
breeding organisms for specific traits (unlike natural selection, where nature dictates survival)
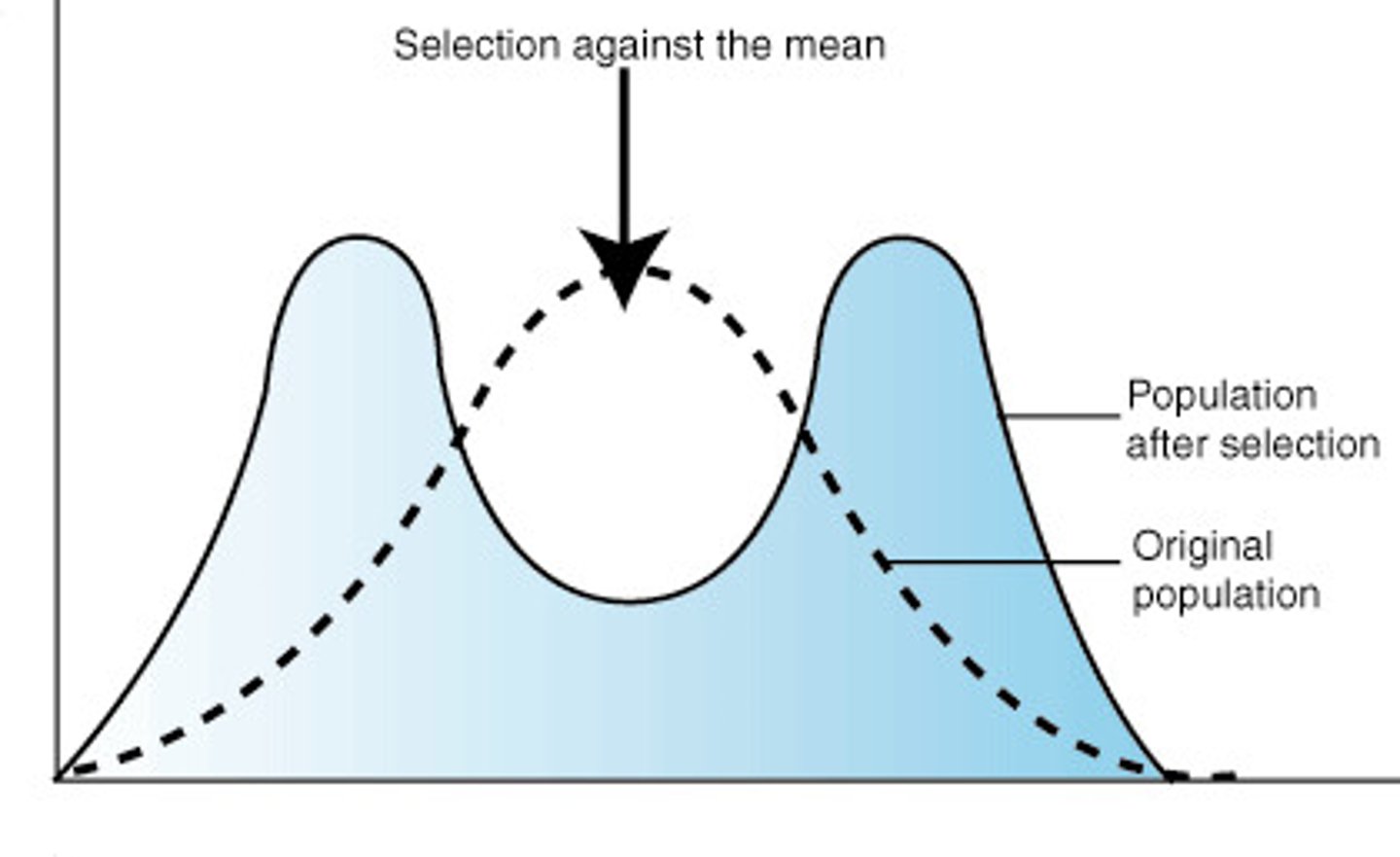
balancing selection
a form of natural selection that maintains genetic variation at a locus within a population
evolutionary trade-off
when a beneficial trait also has a cost, impacting overall fitness
(like bright birds being a target for predators in exchange for more mating chances)

fitness components
life cycle events (survival, mating, reproduction) that affect an organism's overall fitness.
Beneficial mutations always become fixed in a population. (true or false)
False
(factors such as reproduction, death, and random events can prevent beneficial mutations from spreading)

fixed, fixation
when an allele's frequency is 1 (meaning it is the only allele in population, ex: red roses)
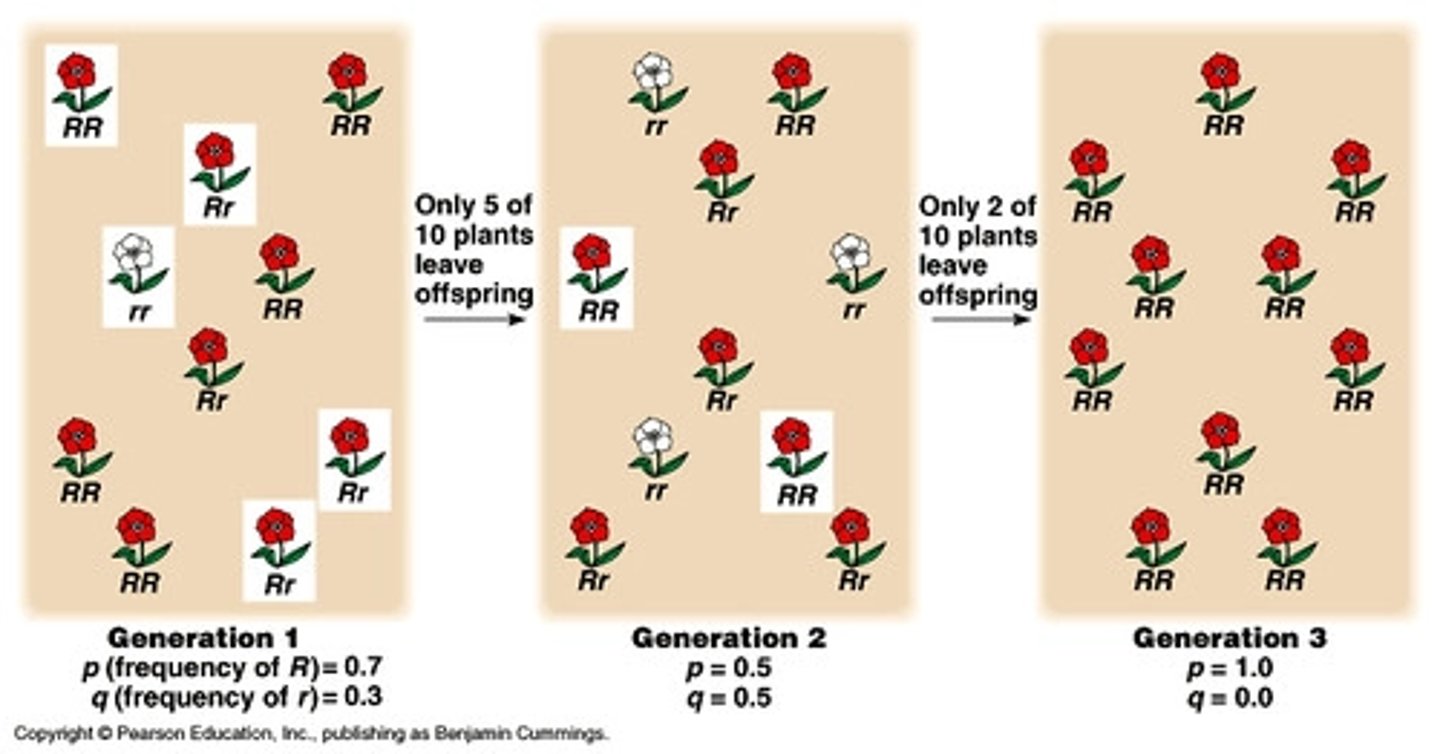
Can positive selection eliminate genetic variation?
Yes, because by the end, only ONE allele remains (in the population), the other is eliminated.
(ex: think of a group with a genotype of all AA).

Does positive selection eliminate mutations?
No, mutations will still occur even if a population is the same (no genetic variation)
(ex: in a population with all AA, aa may pop up due to mutation)
frequency-dependent selection
when a genotype's fitness depends on how common it is in the population

fundamental theory of natural selection
the more variation in fitness exists, the faster the population's overall (mean) fitness improves

genetic correlation
genes influencing multiple traits, causing them to vary together (one cause being pleiotropy)
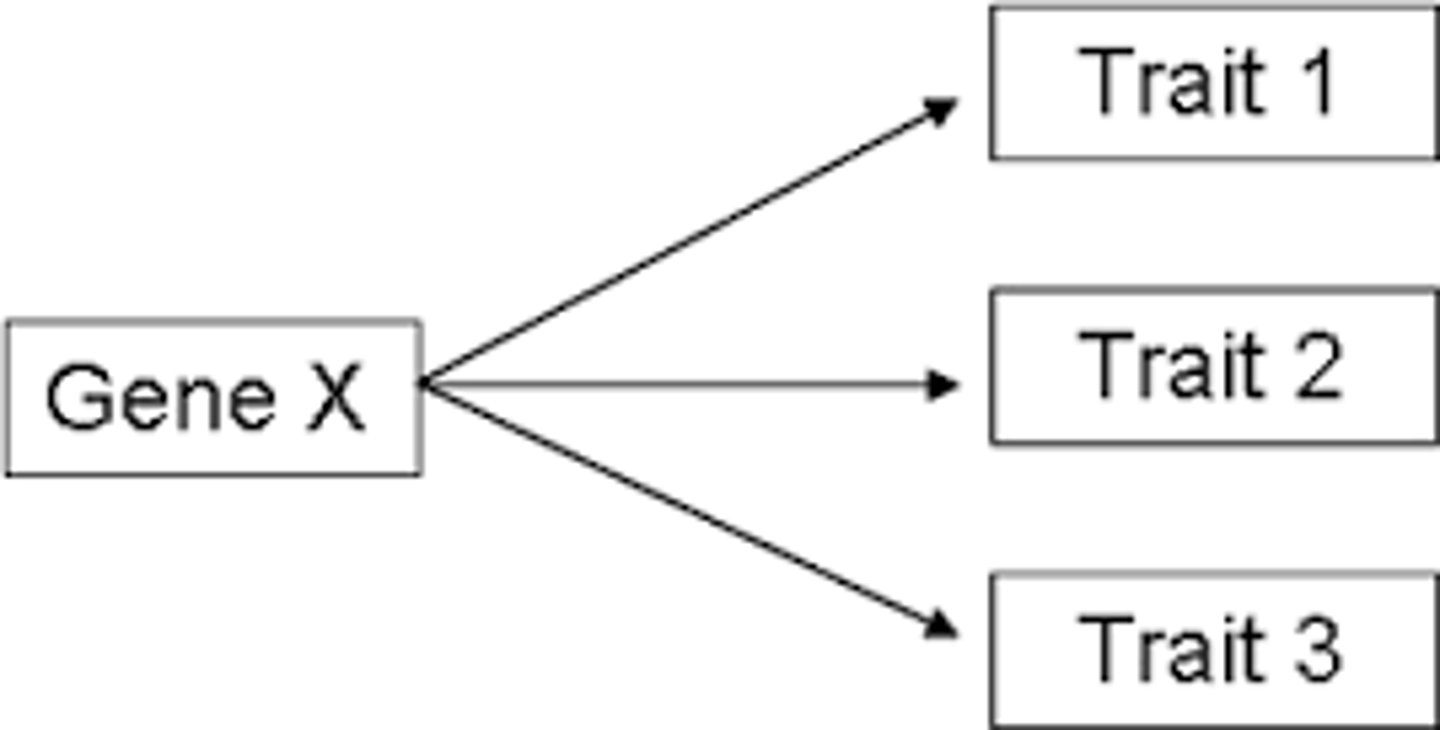
hitchhiking
when the frequency of an allele changes because it's linked to another allele undergoing strong selection
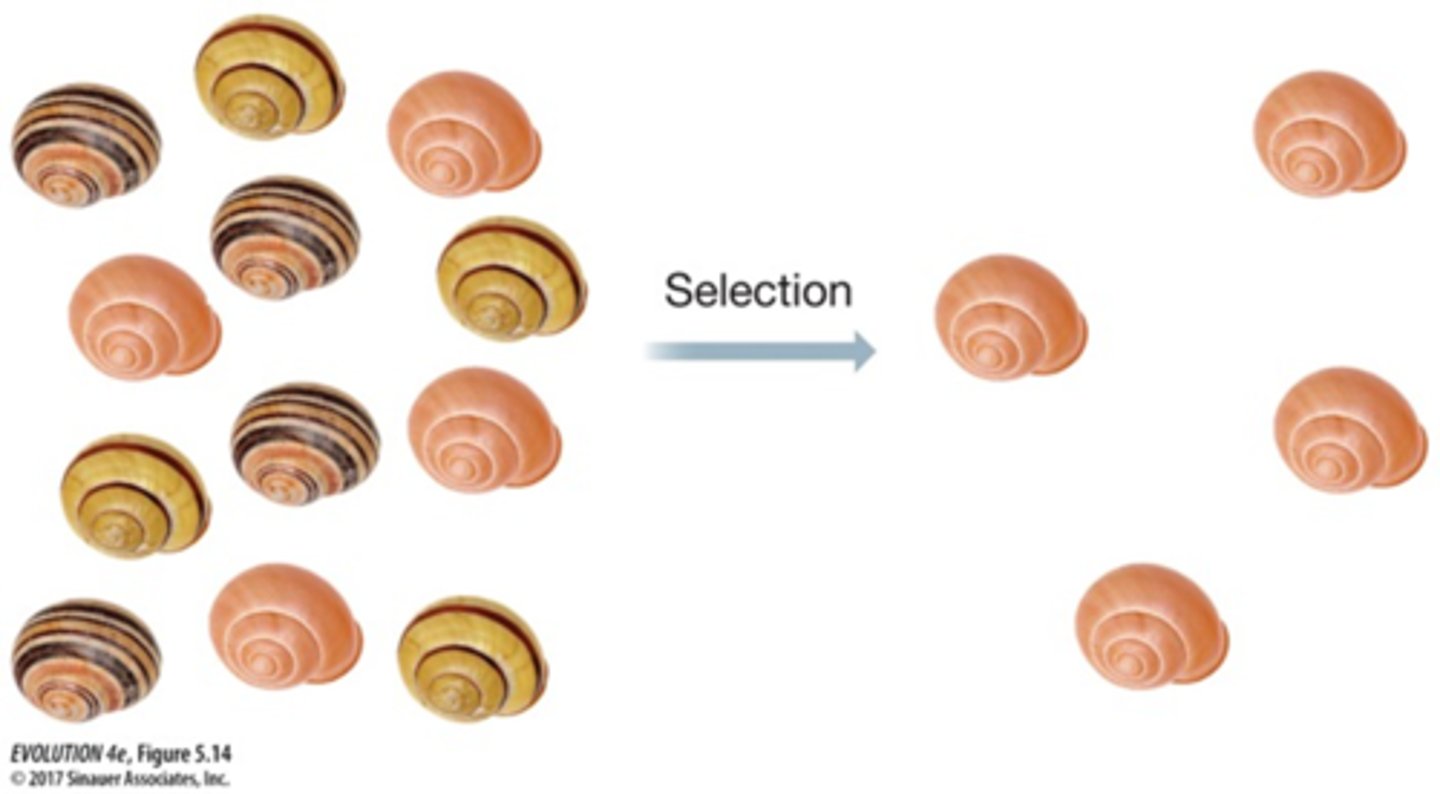
Can hitchhiking occur without linkage disequilibrium?
No, hitchhiking needs linkage disequilibrium (to take effect)
What type of evolution is hitchhiking responsible for?
Evolution of genes that by themselves do NOT affect survival
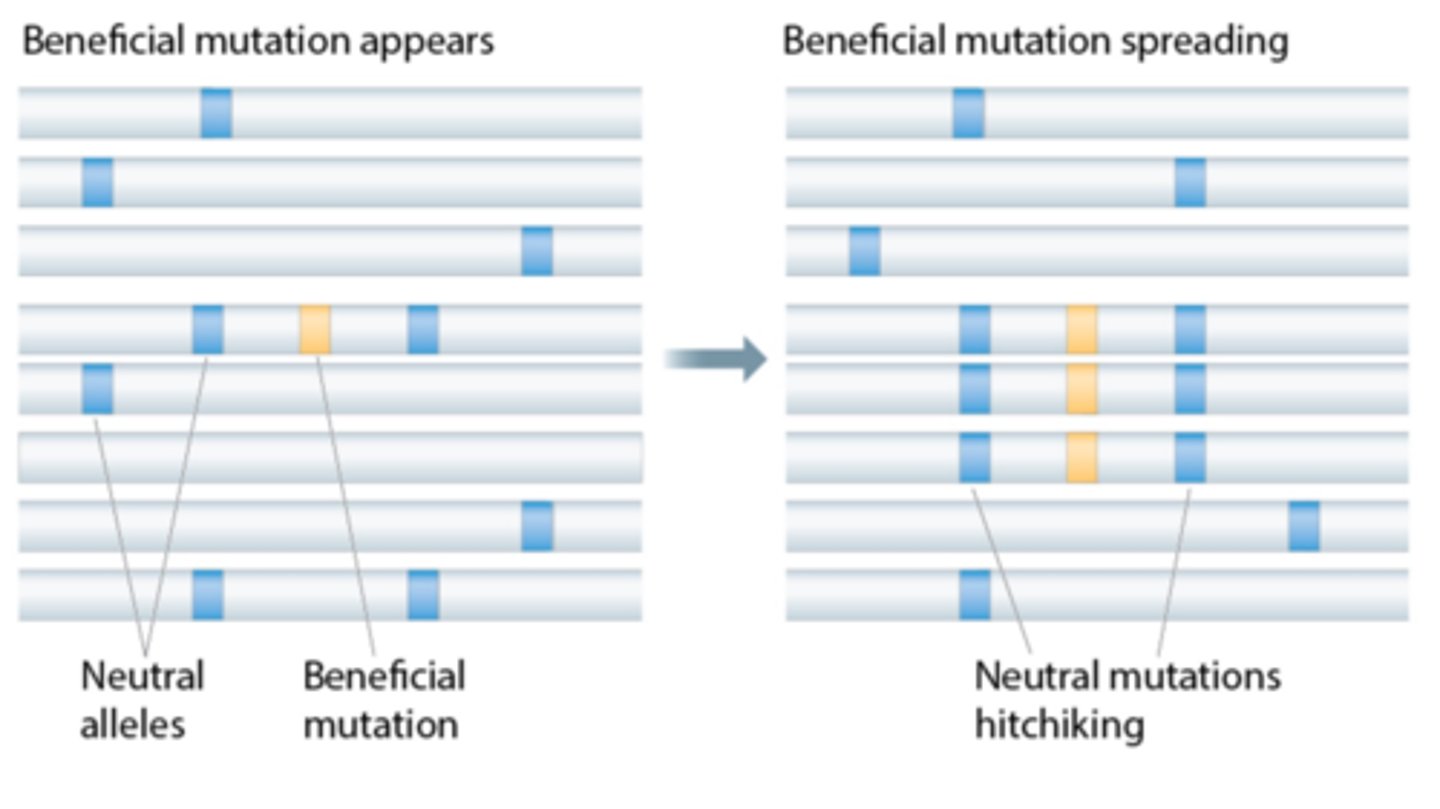
Does standing genetic variation exist before or after selection started?
before
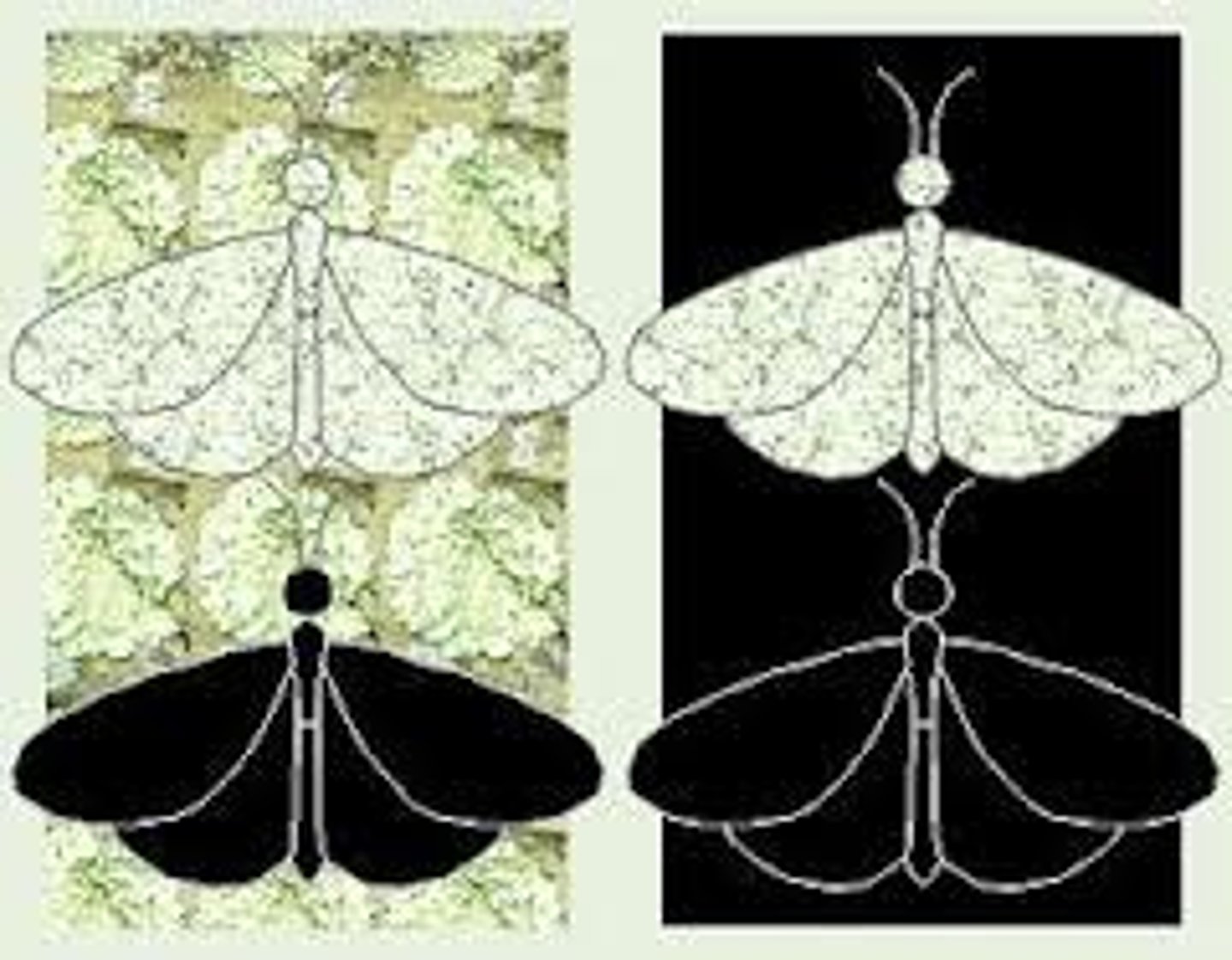
mean fitness
average fitness of all individuals in a population

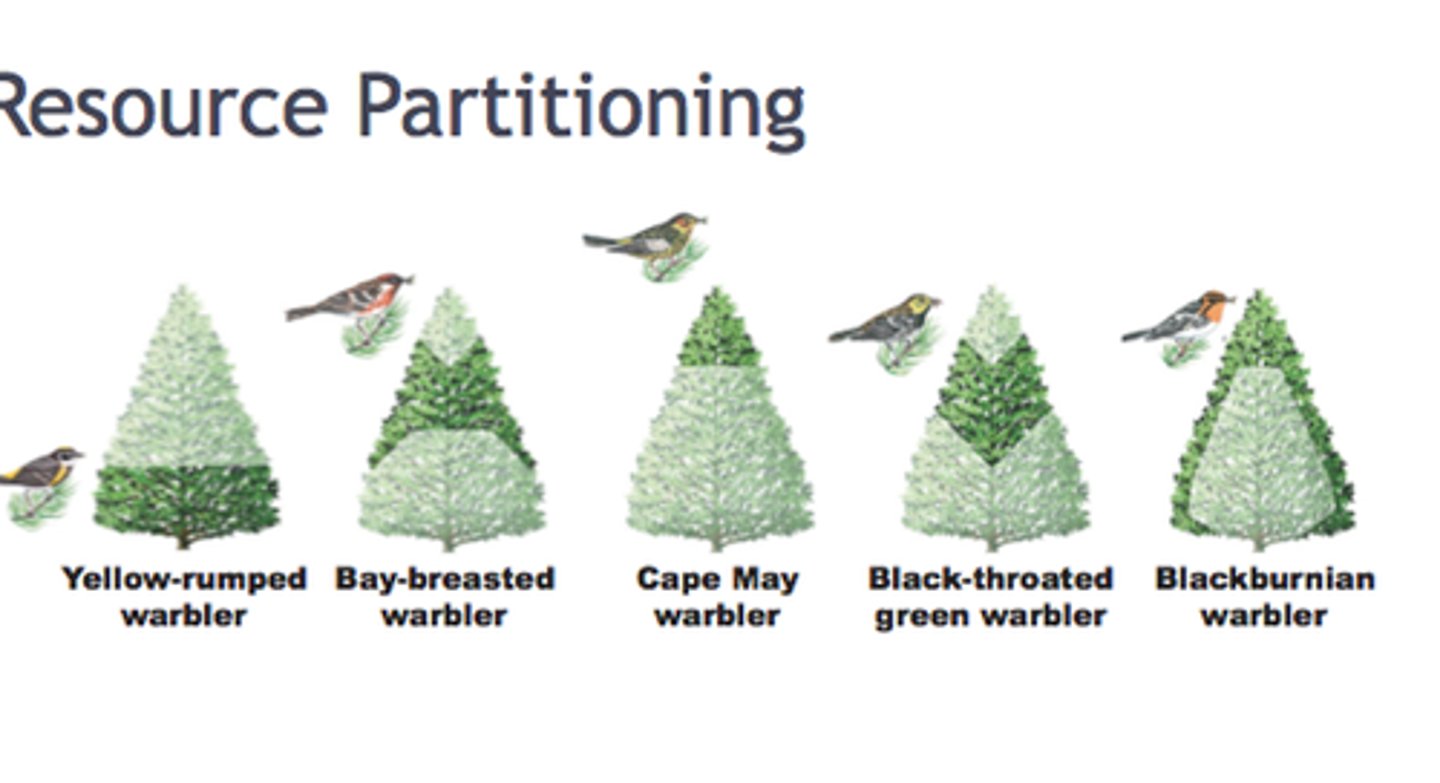
multiple niche polymorphism
when different genotypes thrive in different ecological niches, maintaining genetic diversity
mutation load
proportion of fitness lost due to deleterious mutations

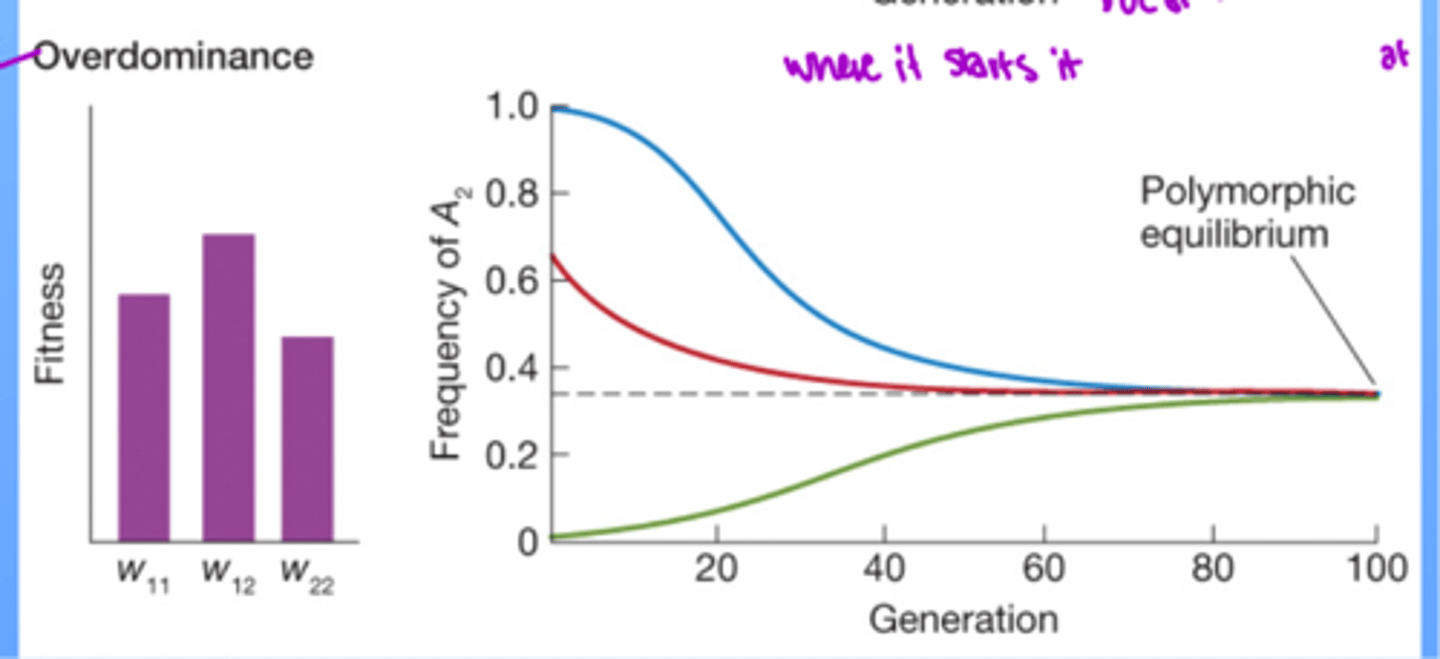
overdominance
when heterozygotes have higher fitness than either homozygote

polymorphic equilibrium
stable state where selection maintains multiple alleles
positive selection
selection favoring alleles that increase fitness
(ex: if dark-furred mice survive and reproduce, dark fur is being positively selected)

purifying selection
elimination of deleterious alleles from a population
relative fitness
the absolute fitness divided by a fitness reference that is agreed on

selection coefficient
shows how strong selection is for a beneficial mutation (how much a genotype's relative fitness differs from a reference)
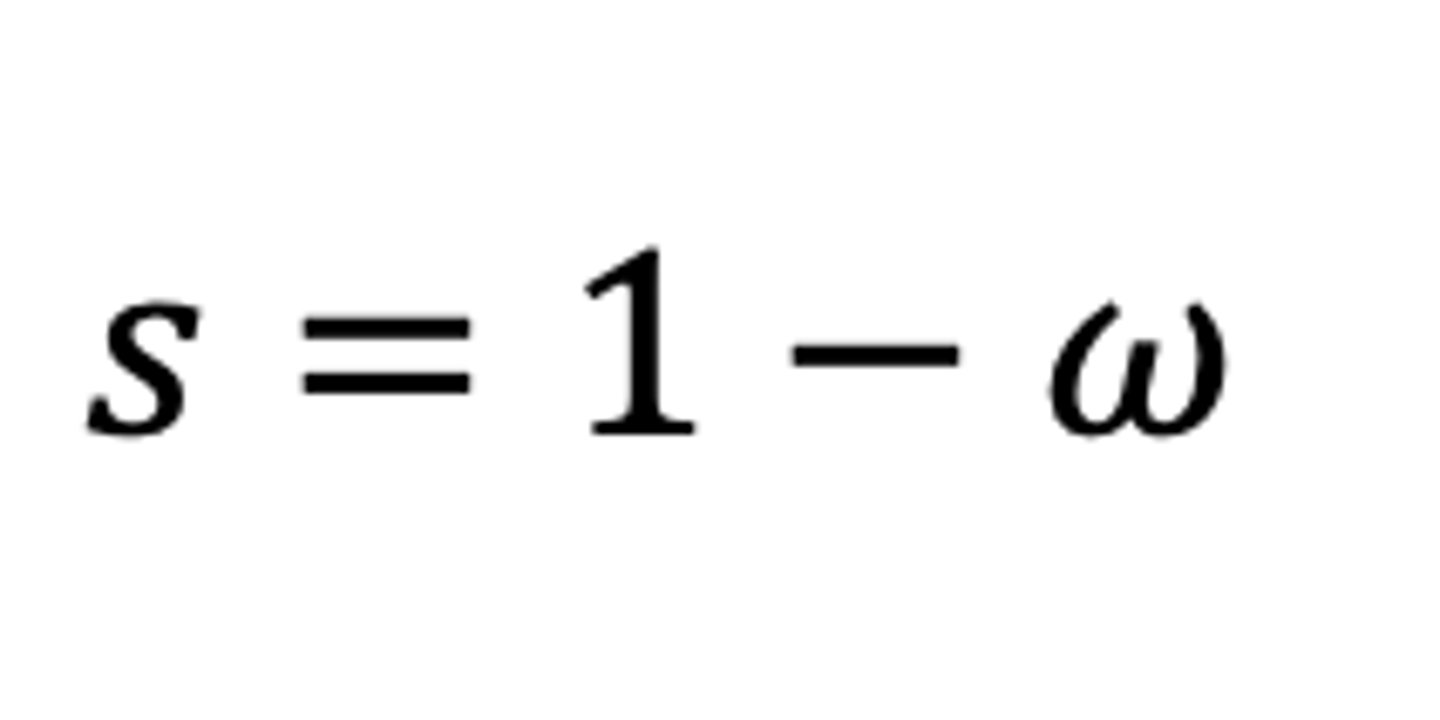
Is the selection coefficient a "fixed property" of an allele?
No (the value of s can change via environment, like pollution and moths)

What affects the selection coefficient?
environment, other alleles in genome
What is the sign of the selection coefficient for a deleterious mutation?
negative
Which is more common, beneficial or deleterious mutations?
Deleterious
If the frequency of an allele A2 is p, what is the frequency of A1?
1-p
(because adding the frequencies of A1 and A2 must equal 1).
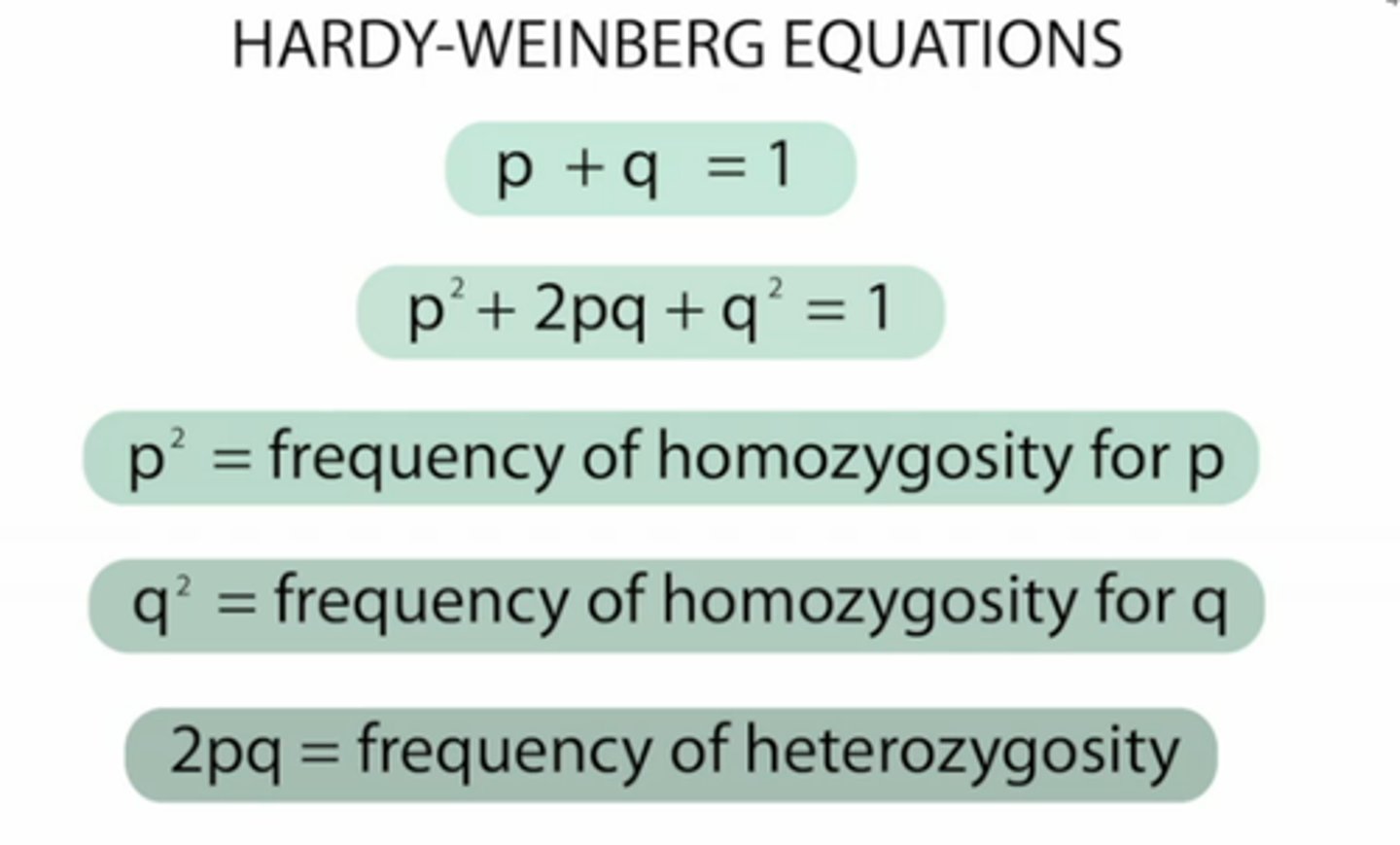
What does it mean when s (selection coefficient) is 0?
No selection is happening.

The rate of evolution is proportional to
strength of selection, amount of genetic variation
selective sweep
rapid spread of a beneficial allele, reducing nearby genetic variation (polymorphism)
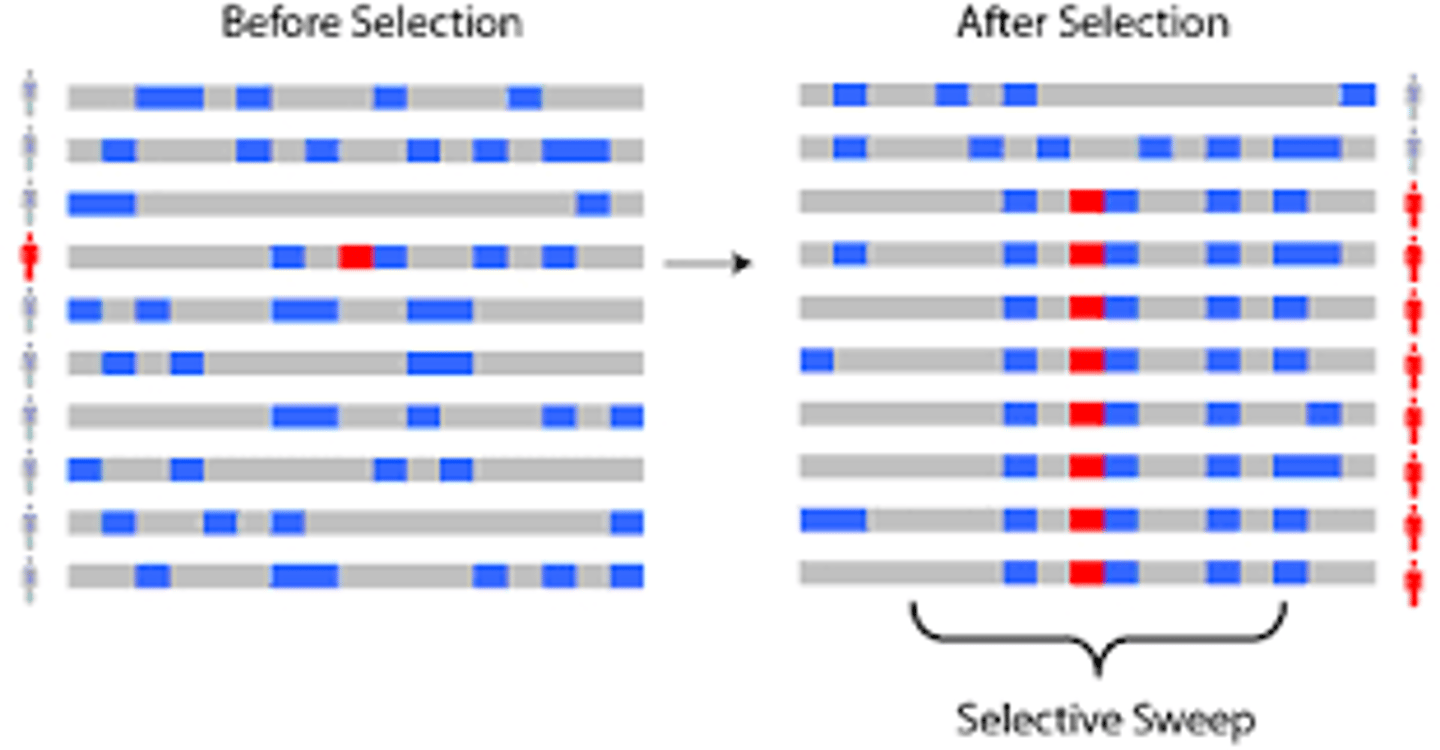
Why are selective sweeps important to geneticists?
Selective sweep reduces genetic variation around a gene, which is a sign that adaptation occurred.
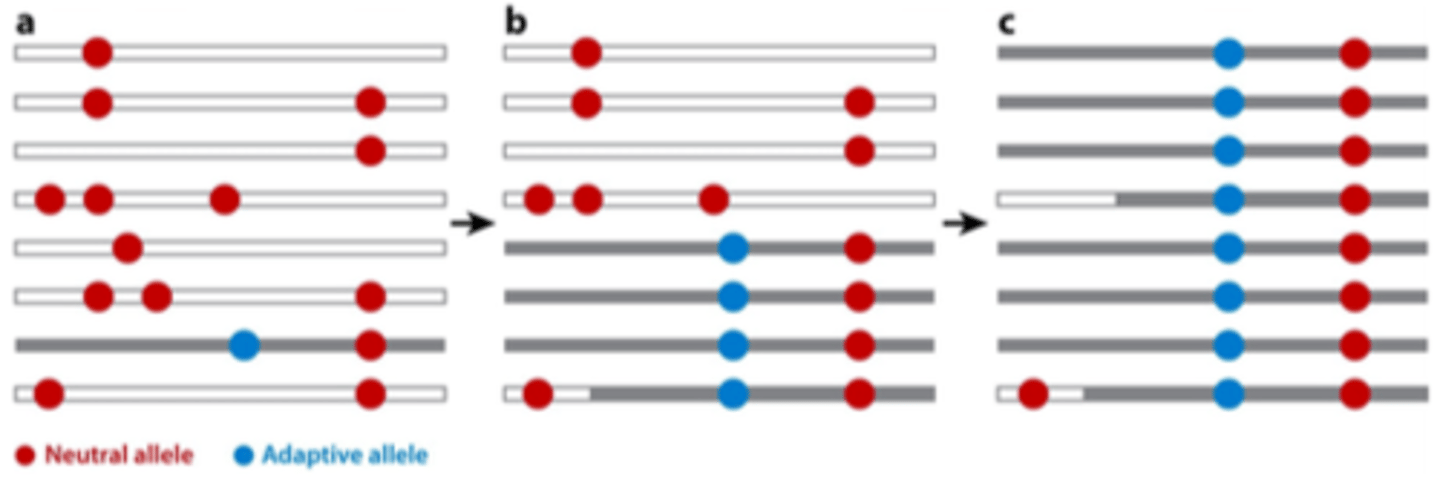
standing genetic variation
when existing alleles (not new mutations) become beneficial due to changing conditions
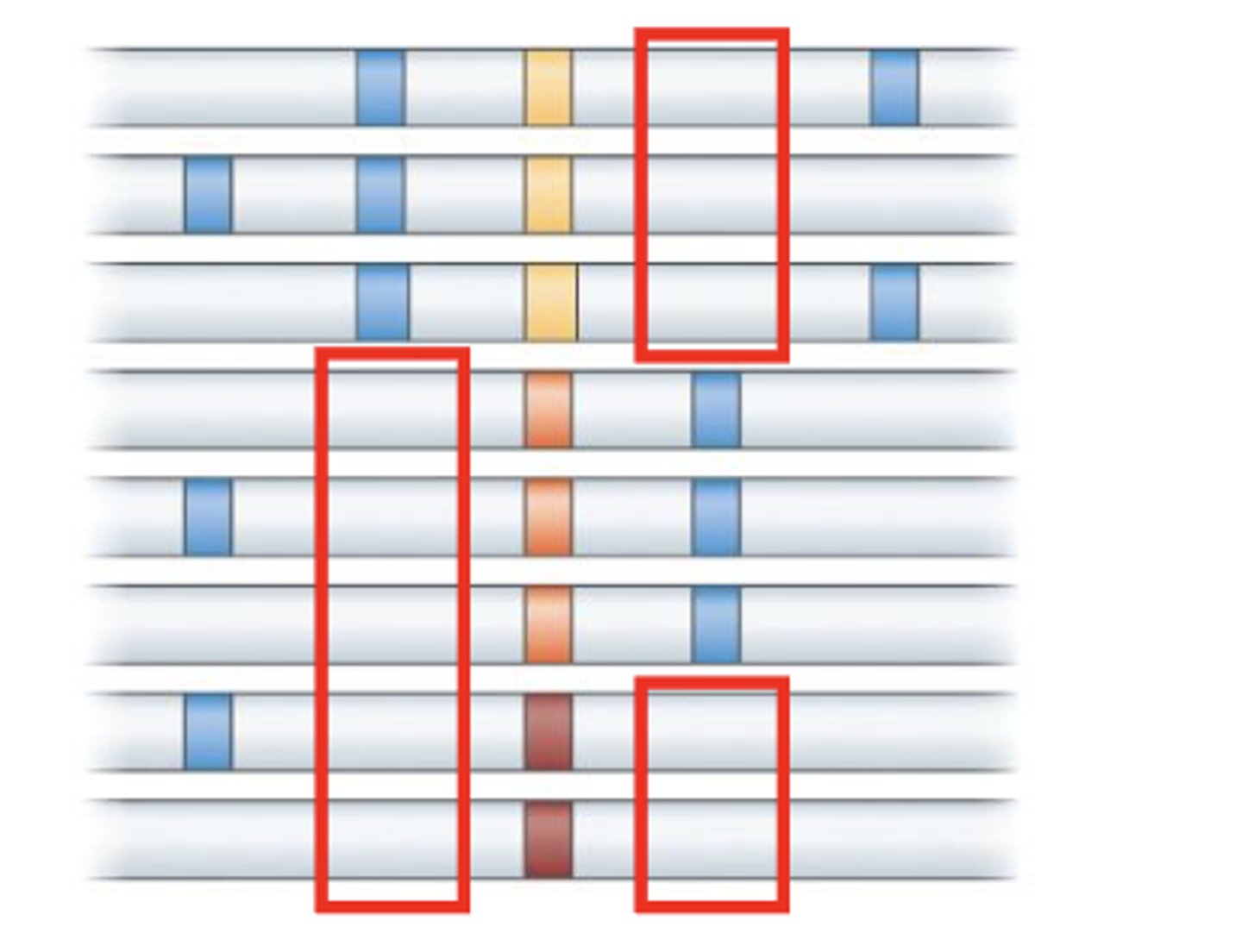
selective sweep vs standing genetic variation
Selective sweep: reduces polymorphism.
Standing genetic variation: acts on existing polymorphism.

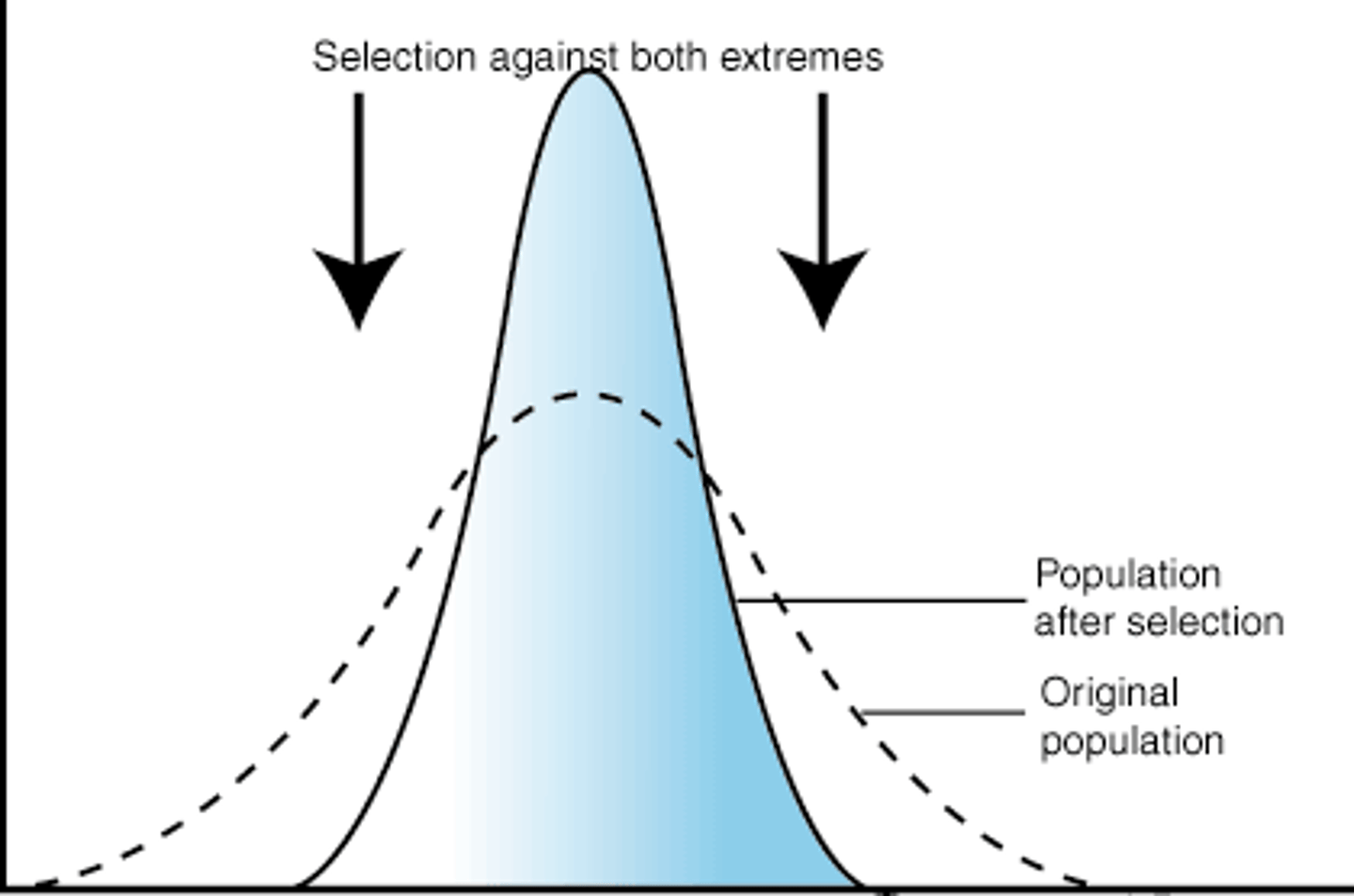
underdominance
when heterozygotes have lower fitness than either homozygotes