BIOL 3000 - Exam 3 - Transcription
1/56
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
57 Terms
Why is transcription likely to occur in G1 or G2?
G1: large amount of enzymes are being prepared for replication
G2: proteins are ready for cell division
Waves of transcription that coincides with different transition points during the cell cycle
Mitosis
Where can transcription NOT occur in the cell cycle?
Template strand
DNA strand that mRNA is built from.
Coding strand
DNA strand that mRNA matches the EXACT sequence of, except mRNA has uracil and this has thymine.
Selective process
Each gene has its own transcription protocol
What is meant by "each gene has its own transcription protocol"?
While most eukaryotes have TATA boxes to initiate transcription, it is not required.
Same for CAAT box and CG box - they may or may not be present in transcription depending on the process.
Transcription always occurs/synthesizes mRNA in the _____'-->_____' direction
5' to 3' (nucleotides are added at the 3' growing tip)
Transcription results in an RNA _________
compliment.
The mRNA is antiparallel and unidirectional to the DNA strand (ONLY ONE STRAND IS MADE INSTEAD OF TWO)
What is the direction of RNA compliment?
Anti-parallel and unidirectional
List the 4 steps of transcription (w/ pre-recognition as 0)
1. recognition
2. initiation
3. elongation
4. termination
Pre-Recognition
During transcription, euchromatin and heterochromatin are packed into the nucleus. If facultative heterochromatin is being used, it must convert to euchromatin prior to transcription. If it stays heterochromatin, it cannot be transcribed.
DNA is packed as ___ and must be turned into ___ before transcription
Heterochromatin, euchromatin
Lys (K) acetylation
Process where either HAT adds acetyl groups to the histone to loosen the DNA from it and prepare it for transcription, or HDAC takes acetyl groups from the histone and tightens the DNA (no transcription)
Histone deacetylase
AKA HDAC. Enzyme that takes away an acetyl group from the histone core. By doing this, the histone tails tighten their grip on DNA and it cannot be transcribed.
Also makes the histone core more positive, which helps the tails grip the DNA tighter.
Step of transcription where transcription factors begin to bind to the promoter region and form PIC; required for RNA polymerase II to bind.
Recognition
TATA Binding protein (TBP)
First transcription factor that has to bind to the promoter region (TATA box); It is a transcription factor ESSENTIAL for transcription.
In recognition, what is recruited by TBP?
General transcription factors
Pre-initiation complex
This is the complex made of general transcription factors, binding to the TBP and the TBP binding to the TATA box. TFIID and TBP BONDING TOGETHER to recruit more transcription factors is what forms this complex.
The whole purpose of this complex is to recruit RNA polymerase.
TFIID
What general transcription factor binds to the TBP to promote more transcription factors and form the pre-initiation complex?
Initiation
Step of transcription where RNA polymerase is recruited to the PIC; RNA polymerase is turned OFF at this stage bc it is unphosphorylated.
Mediator Complex
Complex made up of the PIC and RNA polymerase TOGETHER with RNA polymerase binding to the template strand via its active site.
Brings in ATPase and helicase to unwind DNA and form a transcription bubble.
During initiation, RNA polymerase is __________ at its carboxyl end. In other words, it is turned _____.
Unphosphorylated, off
What process will start the process of RNA polymerase synthesizing mRNA from the template strand?
Phosphorylation (the addition of a phosphate group)
Elongation
Step of transcription where RNA polymerase is phosphorylated at its carboxyl terminal domain (CTD) by the mediator complex.
It goes down the template strand and creates mRNA.
Where and how is RNA Polymerase II phosphorylated?
At its carboxyl end by the meditor complex
The mRNA created during elongation is an exact copy of the coding strand, except for what?
uracils replacing thymines
Termination
Two protein complexes carried by the CTD recognize the poly-a signal (AAUAAA) and terminate the synthesis of mRNA.
The poly-A signal is recognized from the mRNA, NOT the DNA.
What two proteins from CTD carry out termination? What are their functions?
1. Cleavage and polyadenylation specificity factor (CPSF): binds to end of poly-a strand
2. Cleavage stimulation factor (CSTF): cuts the strand to stop synthesis
What is the name of immature mRNA?
hmRNA
Transcription always adds to what prime end?
3'
Immature RNA consists of BOTH ______ and ________.
exons and introns
List 3 steps of post-transcriptional regulation
1. 5' capping
2. 3' polyadenylation
3. RNA processing
5' capping
Co- transcriptional regulation where a guanine group is added to the 5' end of mRNA by capping enzymes found on the CTD; only when 30 nucleotides long
What are the functions of the 5' guanine cap?
Needed for nuclear transport, promotes translation, prevention of mRNA degradation in the cytoplasm, and involved in intron splicing
3' Polyadenylation
Post-transcriptional regulation where a poly(A) polymerase is added to the poly-A site of the mRNA and synthesizes more adenine bases to the 3' end of the mRNA molecule; nascent RNA is cleaved by ribonucelus downstream of conserved AAUAA site.
Functions of Poly A Tail
Enahnce stability of RNA molecule and regulates transport to cytoplasm
RNA processing
Post-transcriptional regulation where introns are removed prior to translation.
Splicing
The mechanism by which introns are removed; large complex of RNA and proteins responsible for removal of introns and transcribed mRNA
Mature mRNA
RNA that is translated to become a polypeptide
Snurp
Small nucelar RNA molecules playing essential roles in splicing
hnRNA
immature single stranded mRNA
Introns
Intervening sequences of RNA that are not expressed in proteins/translated.
Exons
retained sequences of DNA in mature mRNA that are expressed/translated.
Spliceosome
Large complex of RNA and proteins responsible for the removal of introns from transcribed mRNA (splicing); responsible for cleavage of intron and ligation of exons
List the 4 distinct types of introns
1. introns in protein-coding genes, removed by spliceosomes
2. introns in tRNA genes, removed by proteins
3.A. Self-splicing introns which catalyze their own removal
3.B. Self-splicing introns which don't require GTP to remove themselves but do require assistance from proteins
Ribozymes
Self-splicing introns that don't need an enzyme to remove themselves. They do it themselves.
List the ways you define an intron.
1. GU nucleotide sequences at the 5' splice site/donor site
2. AG nucleotide sequence at the 3' splice site/acceptor site
3. polypyrimidine tract (PPT) upstream to the 3' splice site; promotes assembly of the spliceosome
4. branch point sequence (UACUAAC) for snRNP to bind
T/F: A GU and AG sequence at either splice site ALWAYS means you have an intron.
False. These sequences do not determine if you always have an intron, because you must also have the PPT and BPS
List the 9 spliceosome assembly.
1. U1 binds to 5' splice site
2. U2 binds BPS
3. Trimer of U4, U5, and U6 recruited to 5' splice site - U1
4. U1 and U4 dissociation from hnRNA leaving U5 and U6 bound
5. U2 and U6 associate
6. "Lariat" of hnRNA is formed from the intron - U2 and U6 form this by coming together
7. 5' splice site is cleaved and U5 binds to 3' splice site
8. 3' splice site is cleaved and U5 ligates exons together via ATP hydrolysis
9. snRNP's are released along with the spliced intron
What is the most important step of spliceosome assembly?
U5 cuts DNA at two sites AND ligates them back together
Alternative splicing
Regulated process that results in a single gene coding for multiple proteins (aka exon skipping); increases genetic diversity
regulatory molecules, typically proteins, that can interact with cis-acting elements on different chromosomes, enabling flexible and specific gene regulation. Occurs with proteins and RNA.
Trans-acting factors
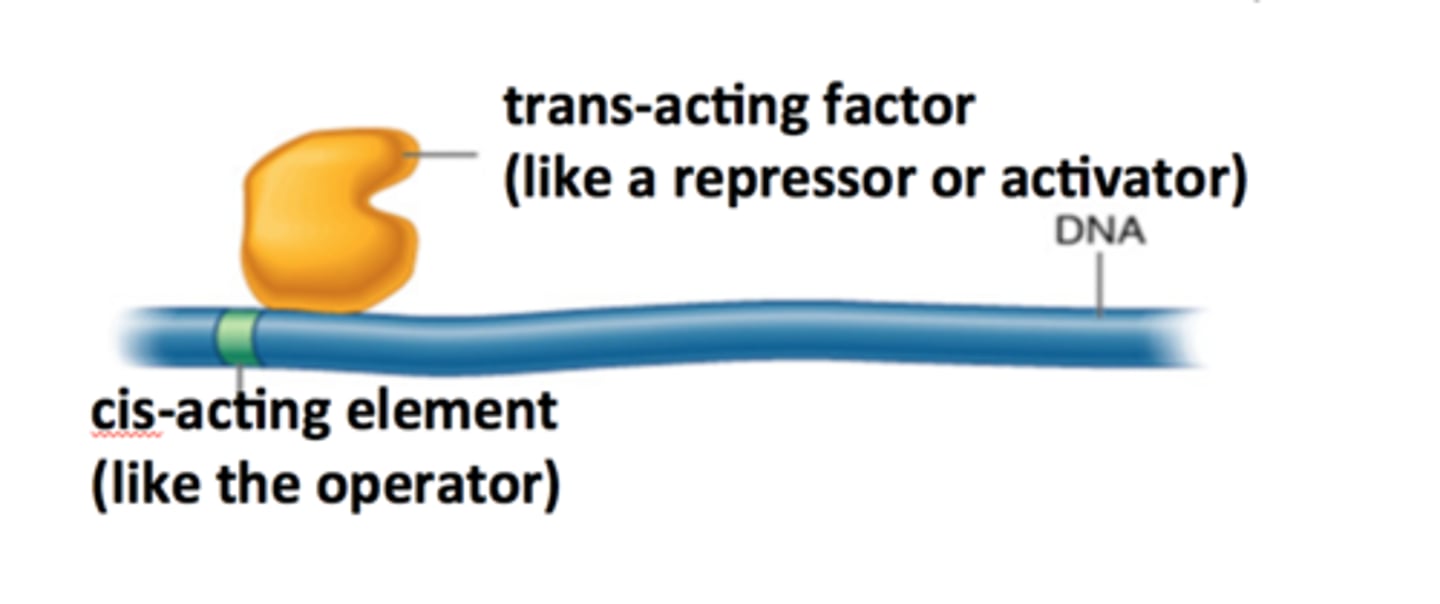
Trans-acting factors
Proteins that control gene expression
Cis-acting factors
DNA sequences in the vicinity of a gene required for gene expression
List the two types of splicing defects.
1. Primary
2. Secondary
Primary splicing defect
Sequences in the mRNA important for splicing are mutated, so no splicing occurs and a defective protein is formed. It is a cis-activating factor.
Secondary splicing defect
Regulatory factors essential for splicing are mutated, so the spliceosome does not know how to splice correct and equal amounts of product are made (trans-activiating factors)