#20: tRNA & tRNA Synthetases
1/62
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
63 Terms
How is the Genetic Code Implemented? → Translation
What was Crick proposing with his “Adaptor Hypothesis”?
the idea of an “adaptor molecule” to bridge RNA chemistry and amino acid chemistry
How is the Genetic Code Implemented? → Translation
What is the adaptor molecule that Crick is proposing with his “Adaptor Hypothesis”?
tRNA
How is the Genetic Code Implemented? → Translation
Single-stranded RNA molecules that have a nucleic acid (the anticodon) on one end and an amino acid on the other
tRNA
How is the Genetic Code Implemented? → Translation
What are the 2 unique components of tRNA?
amino acid, anticodon (the nucleic acid)
How is the Genetic Code Implemented? → Translation
On which end of the tRNA is the amino acid located?
3’
How is the Genetic Code Implemented? → Translation
tRNA forms a series of ______ and ________
stems, loops
How is the Genetic Code Implemented? → Translation
Stems = _______-stranded RNA sections with _____________ (_____-_______ to ________)
double, hybridization, base-paired, itself
How is the Genetic Code Implemented? → Translation
Loops = _______-stranded RNA sections
single
How is the Genetic Code Implemented? → Translation
tRNA is ______ → usually between ___-___ NTPs in length
small, 75-90
How is the Genetic Code Implemented? → Translation
tRNA Secondary & Tertiary Structure
All tRNAs end with the same __ ______ (____) on their ___ end with the ________ being the _________ attachment site for the _______ ______.
3 bases, CCA, 3’, adenine, covalent, amino acid
How is the Genetic Code Implemented? → Translation
tRNA Secondary & Tertiary Structure
tRNAs have a plethora of ___________ _______ throughout the molecule.
modified bases
How is the Genetic Code Implemented? → Translation
tRNA Secondary & Tertiary Structure
All tRNAs have to have a similar ________ structure because they have to fit in the various _________ _____ in the _________. However, this can lead to problems regarding how to achieve this __________ of getting the right ______ _____ to bind to the right ______.
tertiary, binding sites, ribosome, specificity, amino acid, tRNA
How is the Genetic Code Implemented? → Translation
RNA Secondary Structure
When RNA forms a _______ ______ (which is does in the ______ sections of tRNA), it has the same properties of _______.
double helix, stem, A-DNA
How is the Genetic Code Implemented? → Translation
Problem: There are __________ _______ that code for the _____ amino acid. So will each ______ have a different ______ or will the ones that code for the same amino acid have the ______ ______?
Solution: ________!
multiple codons, same, codon, tRNA, same tRNA, wobble
How is the Genetic Code Implemented? → Translation
Wobble
The first ___ bases of a codon _________ the amino acid (these sites are ___________). The ____ position is not as ___________. The ____ base in a codon can “______” and pair with the _____ base in the __________ in non-standard ways. This allows a _______ ______ to bind to multiple _______, thus reducing the # of ______ required to translate all ____ codons.
2, specify, constrained, 3rd, constrained, 3rd, wobble, first, anticodon, single tRNA, codons, tRNAs, 64
How is the Genetic Code Implemented? → Translation
Wobble
What is the “wobble position” in each codon? (1, 2, or 3)
3
How is the Genetic Code Implemented? → Translation
Wobble
What is the “wobble position” in each anticodon? (1, 2, or 3)
1
How is the Genetic Code Implemented? → Translation
Wobble
What are the 3 main rules? (ex: “each tRNA anticodon can either….”)
recognize 1 codon, recognize 2 codons, recognize 3 codons
How is the Genetic Code Implemented? → Translation
Wobble
Describe the situation when a tRNA anticodon recognizes 1 codon
classic base-pairs (C:G, A:U)
How is the Genetic Code Implemented? → Translation
Wobble
Describe the situation when a tRNA anticodon recognizes 2 codons
U can pair with G when it is in the wobble position of either the codon or anticodon
How is the Genetic Code Implemented? → Translation
Wobble
Describe the situation when a tRNA anticodon recognizes 3 codons
inosine in the wobble position of the anticodon can base pair to A, U, or C
How is the Genetic Code Implemented? → Translation
Wobble
When U is in the wobble position of the codon, what can it bind to in the anticodon?
A, G
How is the Genetic Code Implemented? → Translation
Wobble
When U is in the wobble position of the anticodon, what can it bind to in the codon?
A, G
How is the Genetic Code Implemented? → Translation
Wobble
When I is in the wobble position of the anticodon, what can it bind to in the codon?
A, U, C
How is the Genetic Code Implemented? → Translation
Wobble
A common base in tRNA
inosine (I)
How is the Genetic Code Implemented? → Translation
Wobble
The presence of _______ expands the base-pairing capabilities compared with traditional bases
inosine
How is the Genetic Code Implemented? → Translation
Wobble
Inosine is just _____________ __________ (aka a ______________ base + ribose), meaning it can form ___ hydrogen bonds with either ___, ___, or ___
deaminated adenosine, hypoxanthine, 2, A, U, C
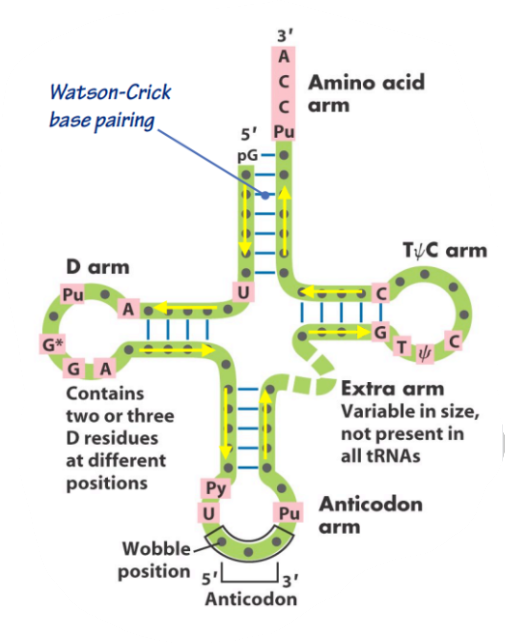
What is this a picture of?
tRNA
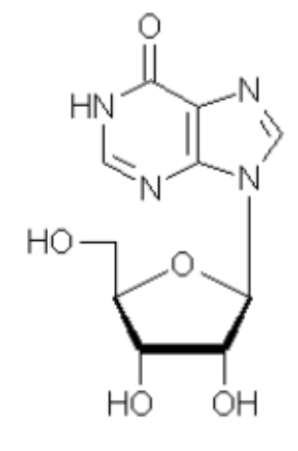
What is this a picture of?
inosine (I)
Translation Parts List → everything we need for protein synthesis
_______ = the template
Amino acids (20)
tRNAs (~ ___)
Aminoacyl tRNA Synthetases (___ → ___ per AA)
_____ + _____
__________ = the enzyme that catalyzes protein synthesis
______ subunit = decoding center
______ subunit = peptidyl transferase center
_________ factors
_________ factors
_________ factors
mRNA, 40, 20, 1, ATP, GTP, Ribosome, small, large, initiation, elongation, termination
Achieving Specificity & Accuracy
The __________ is _____, meaning it adds whichever ______ _____ that is bound to the _____ to the ________ regardless of if it’s the ________ amino acid or not as the ___________ will pair with whichever segment of mRNA it is _________________ to. The __________ does not ____________ the attachment of the ______ _____ to the ______.
Ribosome, blind, amino acid, tRNA, protein, correct, anticodon, complementary, Ribosome, proofread, amino acid, tRNA
Achieving Specificity & Accuracy
The __________ does not ____________ the attachment of the ______ _____ to the ______. This means that the job of getting the ______ amino acid bound and achieving _________ has to be done by the enzyme that puts the amino acid onto the tRNA: the ____________ ______ _______________
Ribosome, proofread, amino acid, tRNA, right, accuracy, aminoacyl tRNA synthetases
Achieving Specificity & Accuracy
Relying on the ___________ to match amino acids to tRNAs is not enough. The various __________ ______ along the tRNA help by providing ________ __________ between the different tRNAs. So even though their _______ has to be similar enough so they all can bind in the __________, the _________ ______ provide enough differences that the __________ _____ __________ (______) can distinguish between the different ______ and can ensure that they place the correct amino acid on each tRNA.
anticodon, modified bases, unique differences, shape, Ribosome, modified bases, aminoacyl tRNA synthetases, aaRS, tRNAs
the enzymes that really know the genetic code and are actually doing the process of translation
aminoacyl tRNA synthetases (aaRS)
The enzyme that places the amino acid onto a tRNA
aminoacyl tRNA synthetases (aaRS)
tRNA Synthetases
There are ___ different types, __ per amino acid.
20, 1
tRNA Synthetases
These are really ____ enzymes, each with very different ___________ from each other and very different ways of _____________ and __________ to their corresponding ______. This is what enables their __________ to ______ the right ______ for their ________ amino acid. They only bind to ______ that carry ___________ that code for their specific amino acid.
big, structures, recognizing, binding, tRNAs, specificity, select, tRNA, specific, tRNAs, anticodons
tRNA Synthetases
There are 2 different groups (______ __ & ______ __) that differ in the type of ___________ that is used to attach the amino acid to its tRNA.
Class I, Class II, chemistry
tRNA Synthetases
What are the 2 main steps for aminoacyl tRNA synthetases adding each amino acid to a tRNA?
activation of amino acid via adenylation, formation of charged tRNA
tRNA Synthetases → Reaction
Step #1: Activation of Amino Acid via Adenylation
Mechanism: An _______ ______ performs a nucleophilic attack on the _____-_________ of an _____. This results in an ___________ _______ ______ (aminoacyl-_____ aka the _________ form). The _________ amino acid forms a _______ __________, meaning there is a phosphoester bond on one side of an oxygen and a regular ester bond on the other end. This also releases ______________, which is cleaved into 2x Pi + heat and ______ the reaction forward.
amino acid, alpha-phosphate, ATP, adenylated amino acid, AMP, activated, acylated, mixed anhydride, pyrophosphate, drives
tRNA Synthetases → Reaction
Step #1: Activation of Amino Acid via Adenylation
Same chemistry as the 1st step of ______ ______ _____-____________
fatty acid beta-oxidation
tRNA Synthetases → Reaction
Step #1: Activation of Amino Acid via Adenylation
Acyl group = ______
R-C=O
tRNA Synthetases → Reaction
Step #2: Formation of Charged tRNA
Mechanism: The ___ ____ (Class I) or ___ ____ (Class II) on the ___-________ on the tRNA attacks the acyl carbon of the __________-_____. This results in the release of _____ and, for Class II aaRS, the aminoacyl-tRNA (_______ _____) final product
Class I aaRS have to go through an additional _______________ step to move the AA from the ___ to ___ position but still eventually reach this same product.
2’ OH, 3’ OH, 3’-Adenine, aminoacyl-AMP, AMP, charged tRNA, transesterification, 2’, 3’
tRNA Synthetases → Reaction
Step #2: Formation of Charged tRNA
_________ ______ = aminoacylated tRNA (tRNA with an attached amino acid)
charged tRNA
tRNA Synthetases → Reaction
Step #2: Formation of Charged tRNA
Each ______ does this a different way, but it doesn’t make much difference in the end as both end with the same ______ ________.
class, final product
tRNA Synthetases → Reaction
Draw the reaction catalyzed by aminoacyl tRNA synthetases (answer in pic)
pic
Translation Errors
What are the 3 main sources of errors in translation?
What can all 3 of these result in?
What can this lead to?
aaRS uses the wrong AA as substrate, aaRS selects the wrong tRNA as substrate, Ribosome selects the wrong AA-tRNA (activated tRNA) for the codon, the wrong AA being put into a protein, major problems like protein misfolding and even neurodegeneration
Translation Errors → Error #1: aaRS uses the wrong AA as substrate
Why would this happen?
Many amino acids are very similar in ________. For example, Valine and Isoleucine only differ in an extra ______ group on Isoleucine. The standard free energy of ___________ _________ is only ~ 12 kJ/mol. This means that, at equilibrium, the active site of Ile RS (Isoleucyl-tRNA synthetase) should contain ~ ___ Valine/____ Isoleucine. This is because (a) ________ can fit in the ___________ binding site and (b) ________ is not all that different. But, the measured misincorporation rate by Ile RS is ____ Valine/______ Isoleucine. So something is increasing _________ (i.e. reducing ______) by >10-fold.
structure, methyl, methylene binding, 1, 200, valine, isoleucine, stability, <1, 3000, accuracy, error
Translation Errors → Error #1: aaRS uses the wrong AA as substrate
How is this avoided?
tRNA synthetases actually have a ______________ mechanism very similar to the “_____” domain in _____ _____________.
EX: If Valine were to bind to the active site of Ile RS, step ___ could still proceed, meaning it would successfully __________ into ______ ___________. However, Ile RS recognizes ___________ Valine as an __________ amino acid and instead of attaching it to the ____________ ______ it ___________ it back into __________ Valine. This results in a ______ _______ of forming phosphodiester bonds and re-breaking them where _____ and _____ are consumed to increase __________.
proofreading, palm, DNA Polymerase, #1, activate, Valine Adenylate, activated, incorrect, isoleucine tRNA, hydrolyzes, inactive, futile cycle, ATP, time, accuracy
Translation Errors → Error #1: aaRS uses the wrong AA as substrate
Various tRNA synthetases do this proofreading in different ways:
Some aaRS proofread the ___________-__________ (ex: Ile RS)
Some aaRS proofread the ___-_______ (so after the AA has been added to the tRNA in step ___)
Some aaRS _____ ___________ at all (ex: the aaRS that puts __________ on because it is a big and bulky molecule that doesn’t really look like many of the other AAs)
aminoacyl-adenylate, aa-tRNA, #2, don’t proofread, tyrosine
Translation Errors → Error #2: aaRS selects the wrong tRNA as substrate
Why would this happen?
tRNAs are very similar in ________. If the ___________ was the only difference between 2 ______ that code for ________ amino acids, this error would be fairly common.
structure, anticodon, tRNAs, different
Translation Errors → Error #2: aaRS selects the wrong tRNA as substrate
How is this avoided?
The various _____ ____________ on the ______ add additional _____________ to each ______ so that the ______ can distinguish the right one to match with its corresponding amino acid.
base modifications, tRNAs, uniqueness, tRNA, aaRS
Which of the following could NOT be used as an anticodon in a cell because it would recognize codons of two different amino acids?
A. (3′) GCI
B. (3′) UCI
C. (3′) AAG
D. (3′) CCC
B
In preparation for attachment to the tRNA, amino acids are activated by…
A. adenylation
B. N-acetylation
C. formylation
D. methylation
A
How many amino acids are there in the standard genetic code?
A. 4
B. 20
C. 16
D. 64
E. 3
B
Which of the following is NOT a feature of tRNA molecules?
A. They are linked to amino acids at their 3' end.
B. About half the nucleotides in tRNA are base-paired.
C. They contain many unusual bases.
D. Each is a single chain between 73 and 93 ribonucleotides.
E. They are synthesized by RNA polymerase II.
E
What does it mean when the genetic code is described as "degenerate?"
A. It means that the translation machinery is prone to making errors.
B. It means that more than one codon can specify the same amino acid.
C. It means that there are fewer codons than amino acids.
D. It means that two or more anticodons can base pair with the same codon.
B
Which of the following is true about aminoacyl-tRNA synthetases?
A. They cannot proofread.
B. They recognize only the anticodon arm of tRNAs.
C. They add an amino acid to the 5′ end of a tRNA.
D. They generate charged tRNAs.
D
What is the anticodon on a tRNA that carries Trp, written 5’ → 3’?
A. CCA
B. ACC
C. UGG
D. GGU
E. None of the above
A
Which of the following would be a suitable anticodon for the valine tRNA?
A. 5’-CAI-3’
B. 5’-IAC-3’
C. 5’-IGC-3’
D. 5’-IAU-3’
E. None of the above
B
Which of the following best characterizes the relationship between amino acids and tRNAs?
A. The activation of an amino acid by formation of an aminoacyl-tRNA is coupled to the hydrolysis of ATP to AMP + 2pi.
B. The conformation of an aminoacyl-tRNA facilitates the direct interaction between the amino acid and its appropriate codon in the mRNA-ribosome complex.
C. Formation of the ester linkages between a tRNA and its corresponding amino acid is catalyzed by the tRNA itself.
D. A tRNA binds to its appropriate amino acid through a covalent linkage of the amino acid’s side chain to the base of the nucleotide immediately 5’ of the anticodon.
E. A tRNA is a six-nucleotide RNA molecule consisting of an anticodon followed by a CCA sequence that accepts amino acids.
A
Which ONE of the following statements about tRNA is FALSE?
A. A tRNA may recognize more than one codon.
B. All tRNA have a CCA sequence at their 3’ end which is the site of aminoacylation.
C. tRNAs can have thymine in their sequence.
D. An amino acid is added to the end of the tRNA by the nucleophilic attack of the amino acid carboxyl group on the terminal phosphate of the tRNA.
E. All of the above statements are true.
D
Which best explains how the valine aaRS is prevented from incorrectly adding isoleucine to the valine tRNA?
A. An isoleucine adenylate would be hydrolyzed at the synthesis site of the valine aaRS.
B. An isoleucine adenylate would be hydrolyzed at the editing site of the valine aaRS.
C. A valine adenylate would be hydrolyzed at the editing site of the valine aRS.
D. Isoleucine is too big to bind to the synthesis site of the valine aaRS.
E. Isoleucine is too big to bind to the editing site of the valine aaRS.
D