Genetics/BIOL 3000 (Seibenheiner) - Exam 3, BIOL 3000 EXAM 3
1/474
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
475 Terms
23 pairs, 46 chromosomes
How many PAIRS of homologous chromosomes must fit inside a nucleus? How many TOTAL chromsomes?
6 billion
What is the estimated amount of base pairs possible?
2 meters (6.5 ft)
There is approx. _____ meters of DNA packed inside one cell.
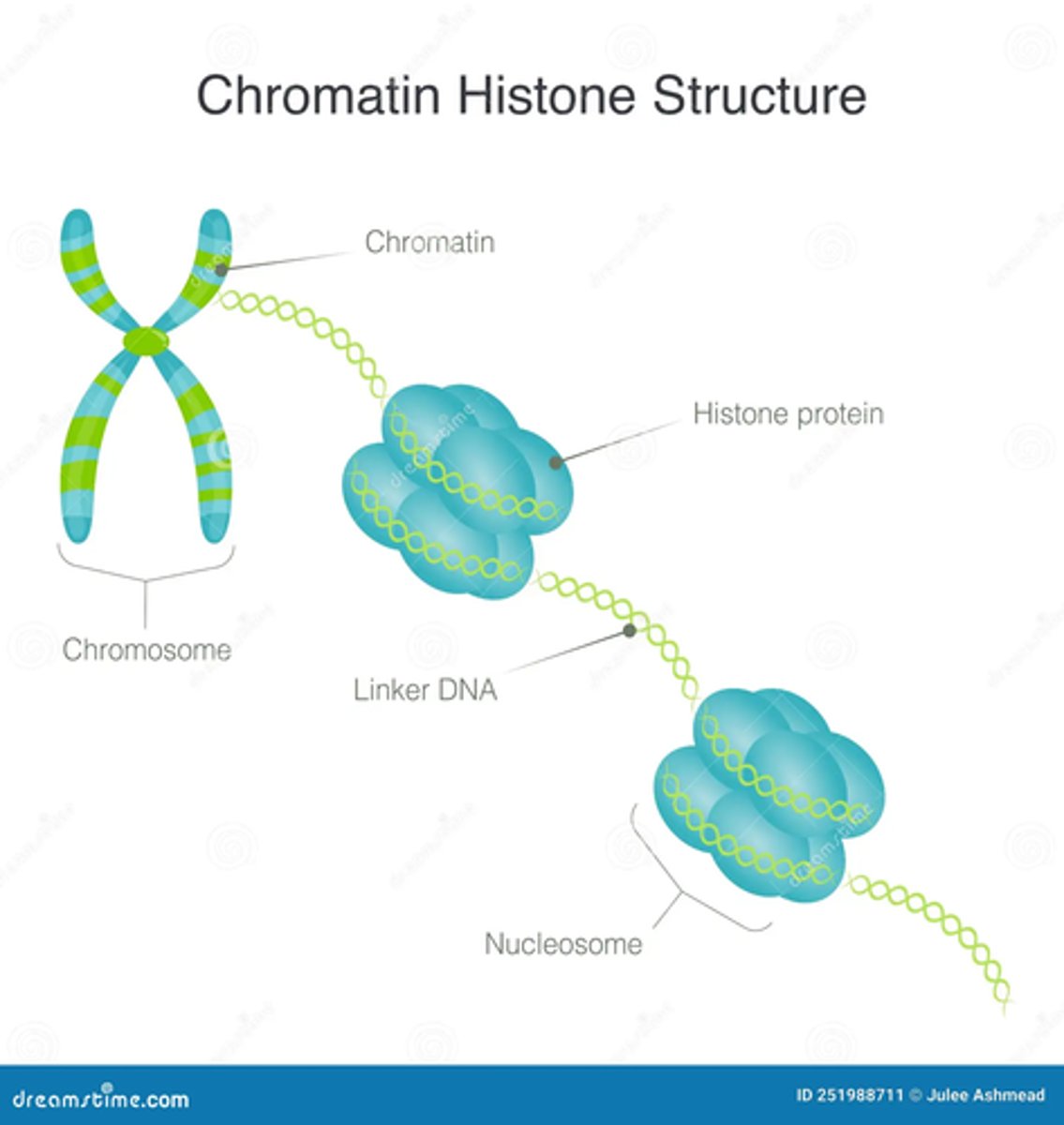
Chromatin
A combination of DNA and proteins (histones) inside the cell
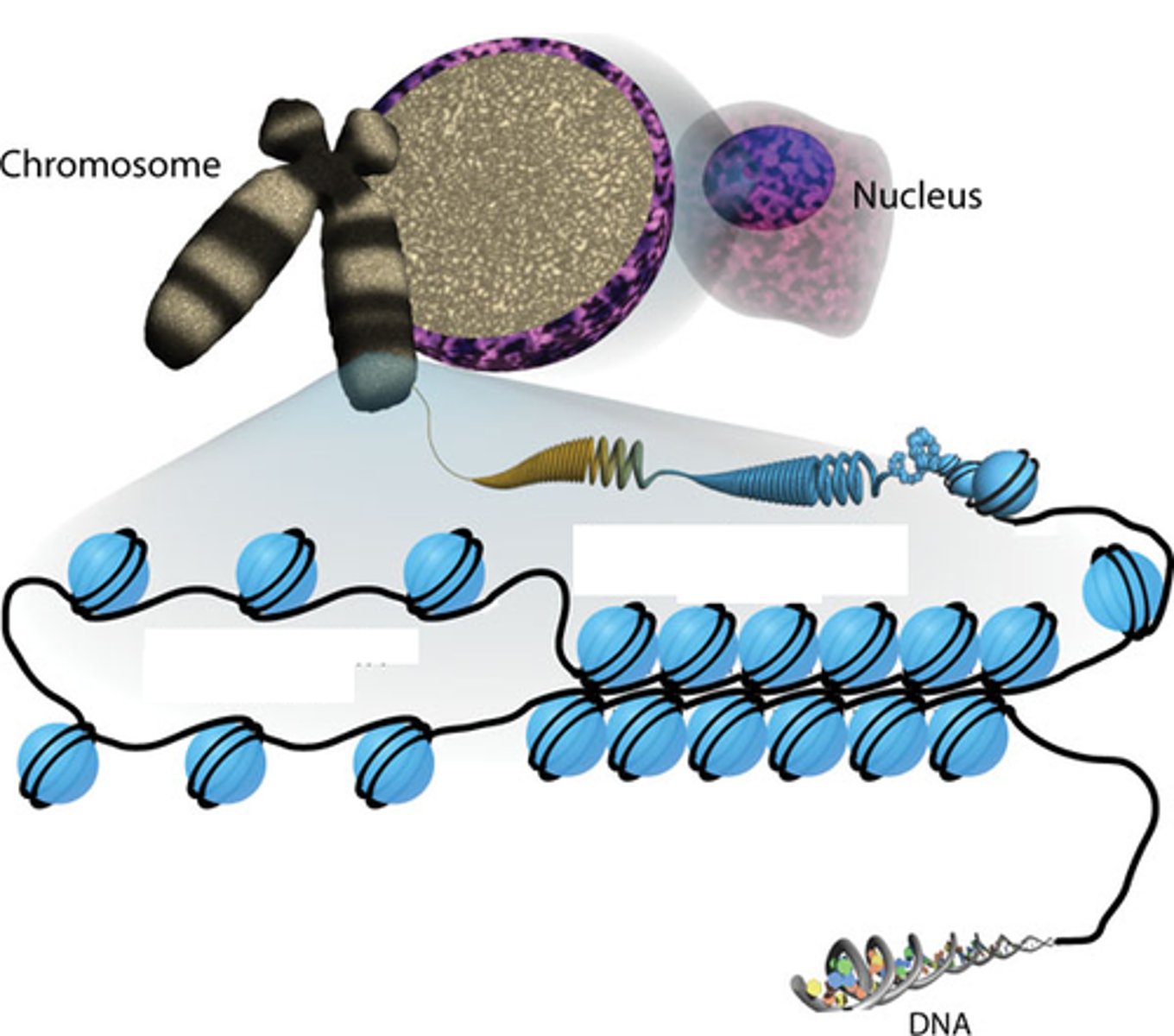
Histones
type of protein specifically involved in forming chromatin
1. euchromatin
2. heterochromatin
List the two types of chromatin structures present within a chromosome.
Euchromatin
Type of "active" chromatin with a loose structure. It is the most active chromatin pattern out of the two types because the DNA is easily accessible/readable by proteins and enzymes (ex. RNA polymerase).
Heterochromatin
Type of "silent" chromatin pattern with a tightly packed structure and genetically inactive sequences. Enzymes and proteins cannot access the DNA because of its tightly packed nature.
1. constitutive heterochromatin
2. facultative heterochromatin
List the two types of heterochromatin.
constitutive heterochromatin
Type of heterochromatin where the centromere is ALWAYS heterochromatic (tightly packed), therefore it cannot be accessed/read at ALL
facultative heterochromatin
Type of heterochromatin where the centromere can change between being heterochromatic (tightly packed) and euchromatic (loosely packed), so it can SOMETIMES be read (only when it is in a euchromatic state).
1. Folded Fiber Model
2. Nucleosome Model
Which two models have been used to explain the association of DNA and protein (AKA chromatin as a whole)?
folded fiber model
Early model (incomplete) used to explain chromatin. Proposed that there were whole mounts of white blood cells. It found few or no free fiber ends and concluded that each chromatid must consist of a single fiber (like a ball of string). Said that replication occurred from each end toward the centromere.
Proposed Type A and Type B DNA fibers.
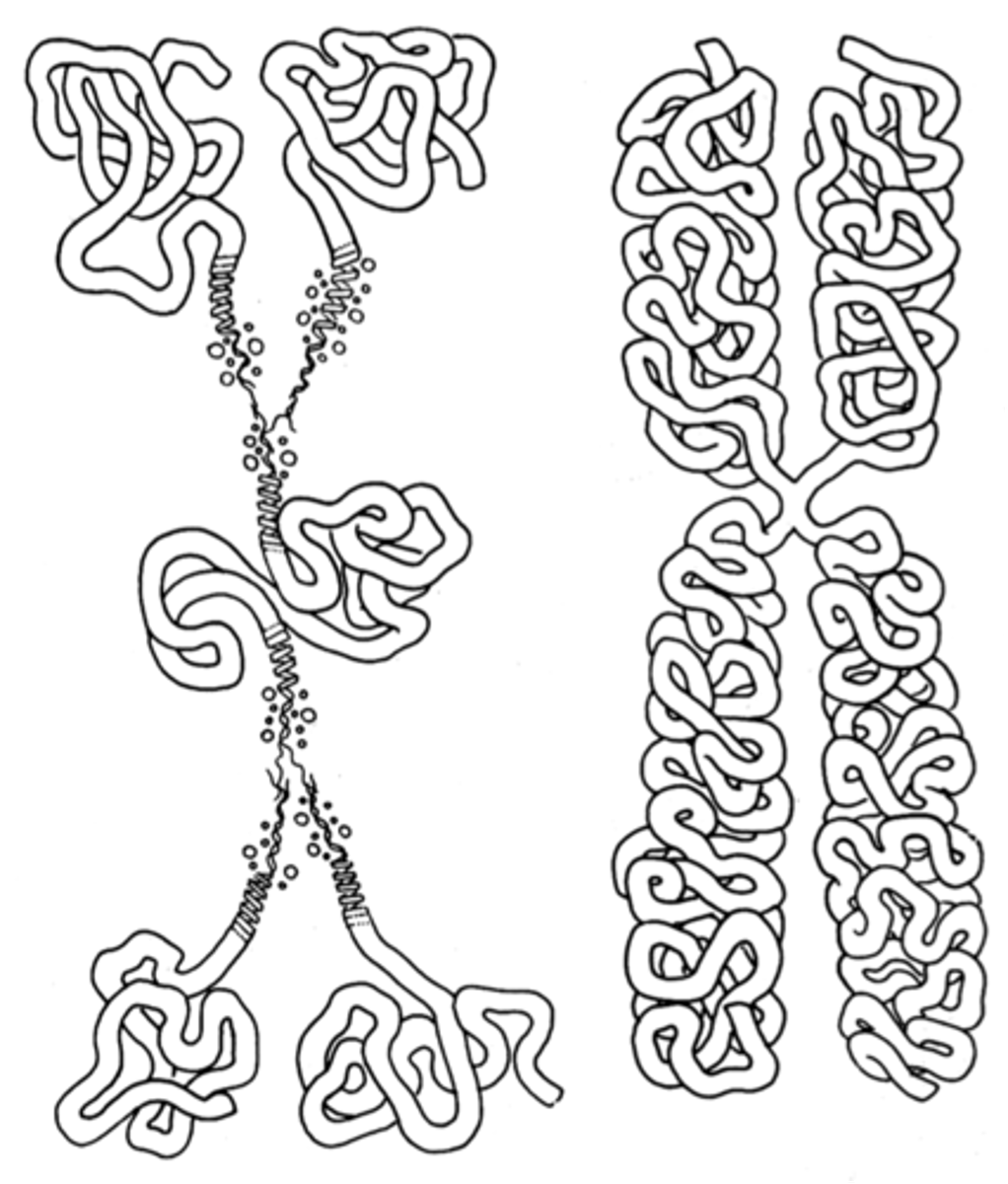
Type A fiber
Type of DNA fiber in the folded fiber model that was said to be 1-10 nm (ratio of 6:1)
Type B fiber
Type of DNA fiber in the folded fiber model that was said to be 20-25 nm (ratio of 10:1)
Type B
The folded fiber model said that extensive folding of Type ___ formed a chromatid.
Nucleosome Model
Most commonly accepted model for DNA packaging. Proposed multiple levels of compaction, with DNA being wrapped around histones to form a structure known as a nucleosome.
Localized areas of transcription and was an overall better fit for protein biosynthesis.
Nucleosome
Type of structure made of DNA wrapping around core histones (proteins)
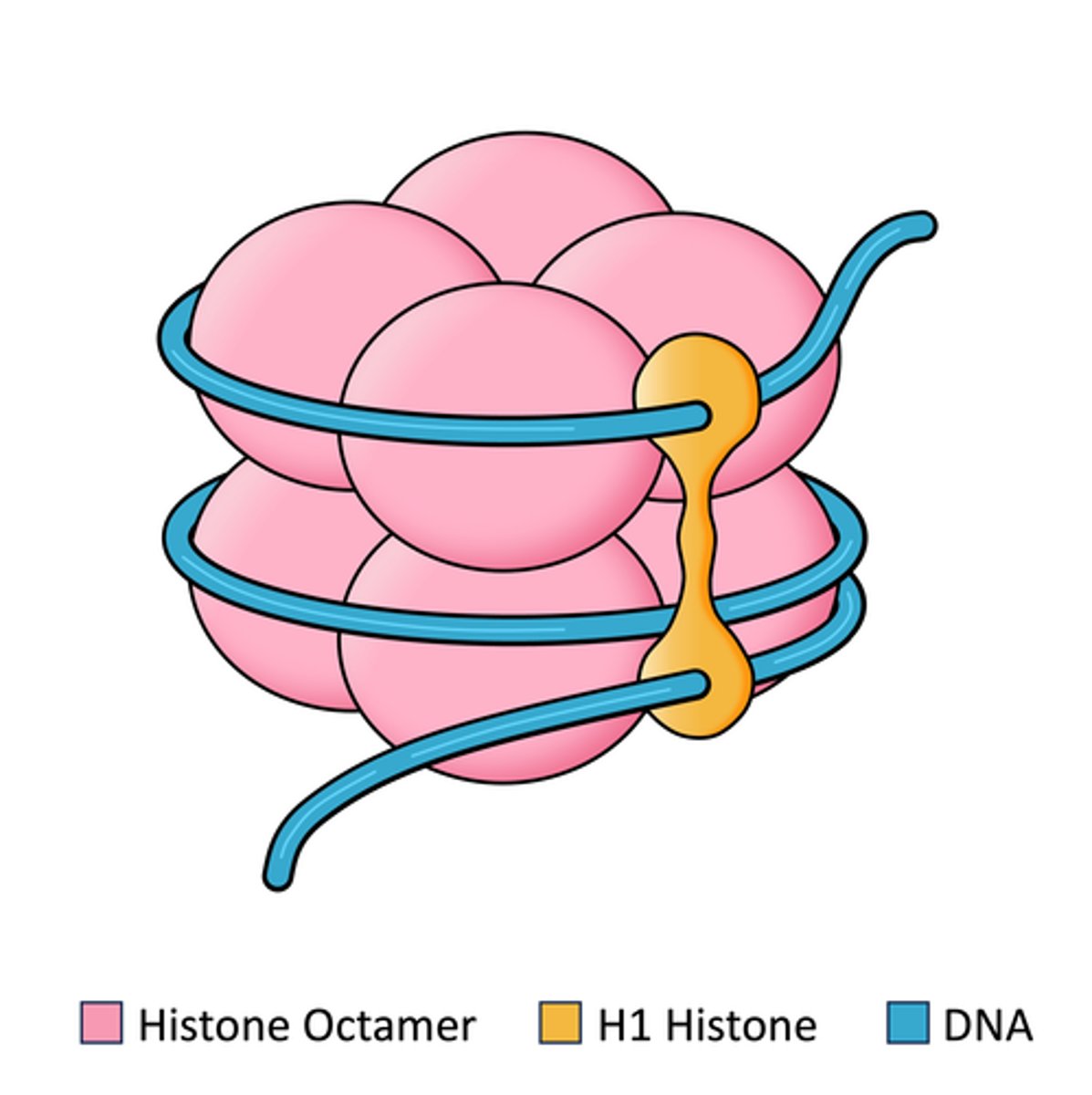
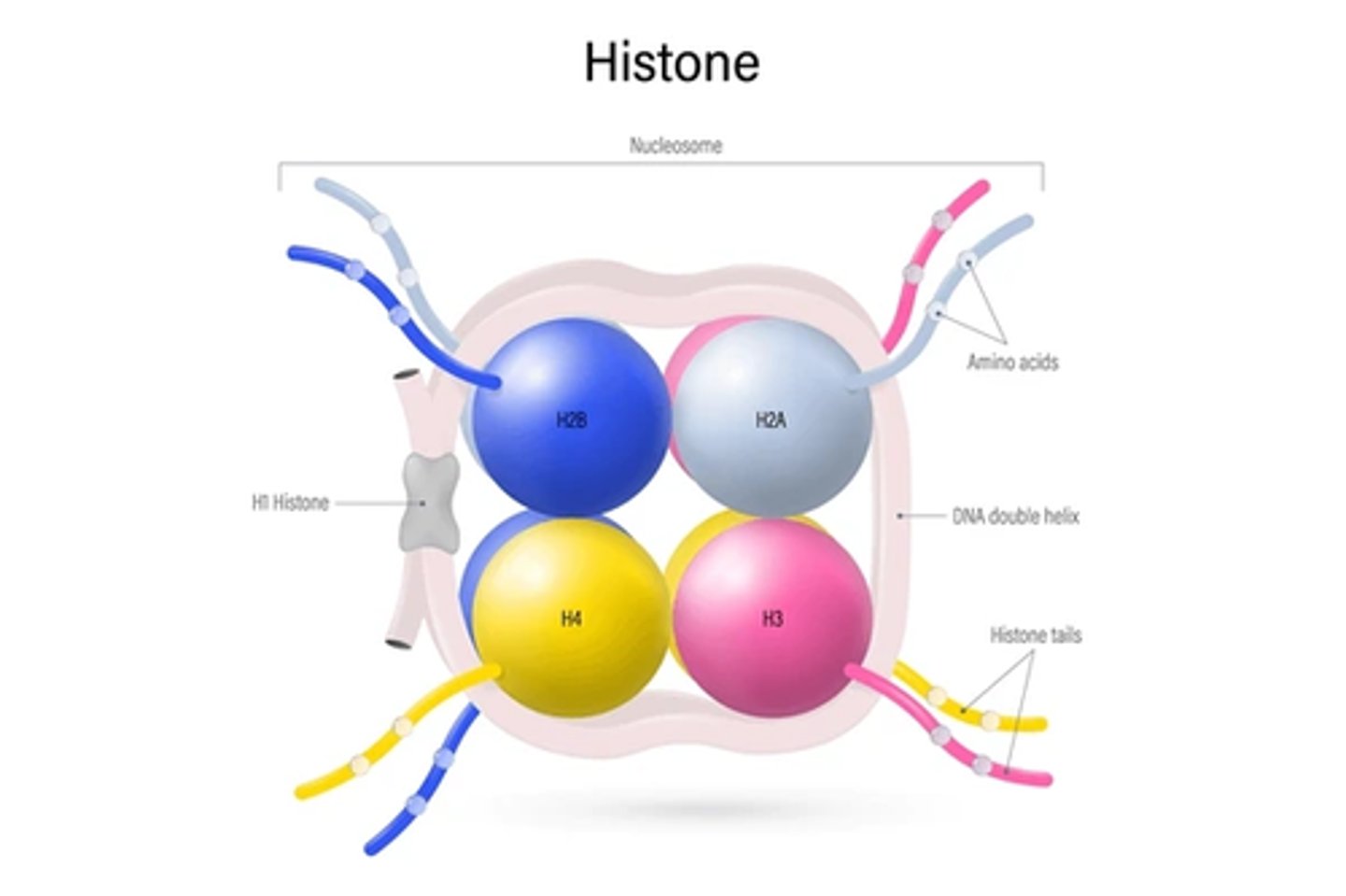
1. core histones
2. Linker histones
List the two classes of histones
Histone core
Histone cluster with DNA (1.5 turns) around it. Made of core histones and linker histones together
positively, negatively
The histone core is very ________ charged, while DNA is very ________ charged.
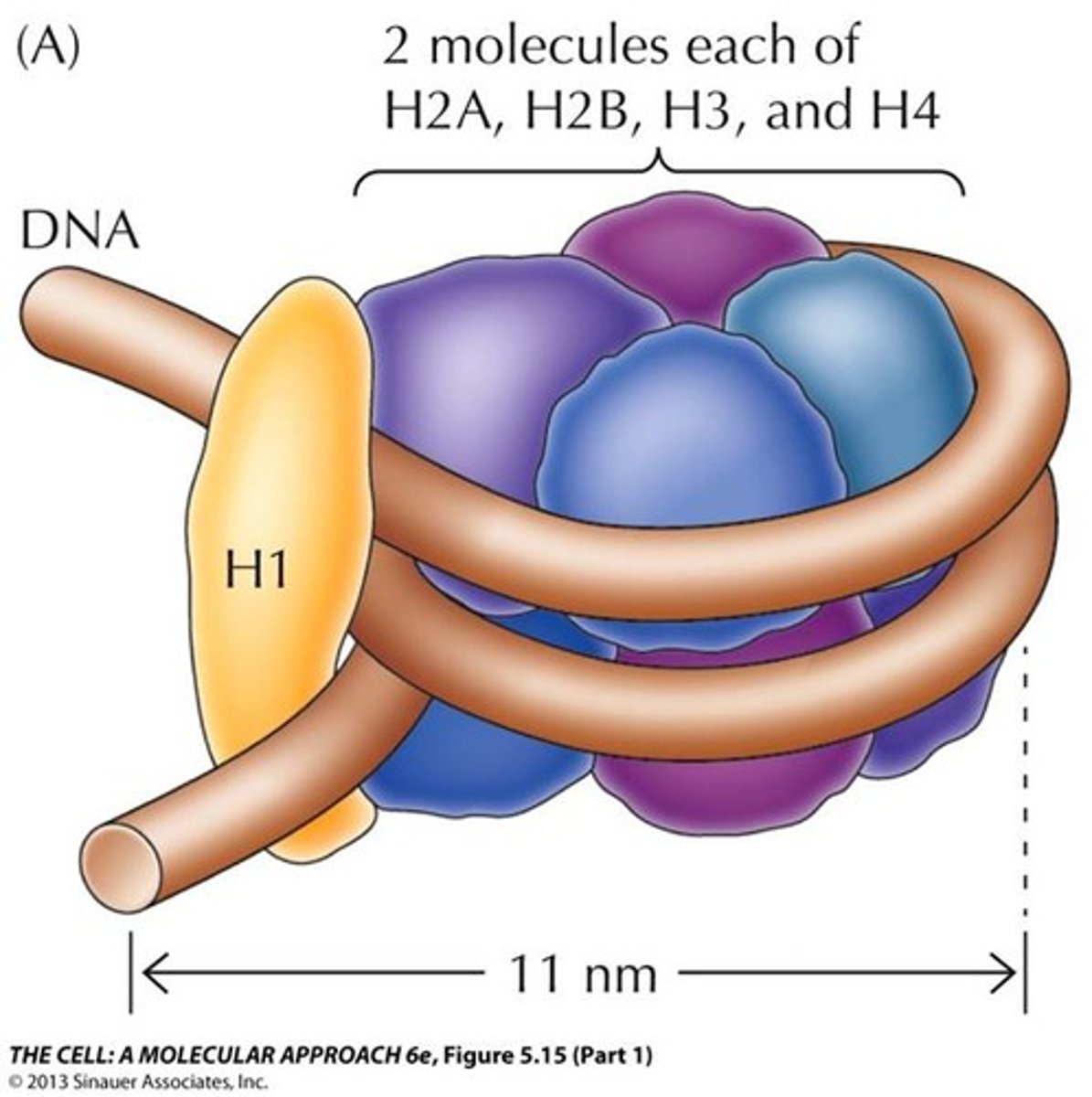
Core histones
Class of histones consisting of 8 total proteins.
Consists of ~120 amino acids. Highly conserved during evolution. Combine to form the histone core. VERY BASIC in charge (approx. 25% lysine and arginine).
Core histone. Called this bc H2A, H2B, H3 and H4 are always turned on and necessary for properly compacting DNA
Which histone is referred to as a "housekeeping protein" and why?
They always have the same sequence and structure throughout the body
What is meant by "core histones are highly conserved during evolution"?
1. H2A
2. H2B
3. H3
4. H4
List the core histone proteins.
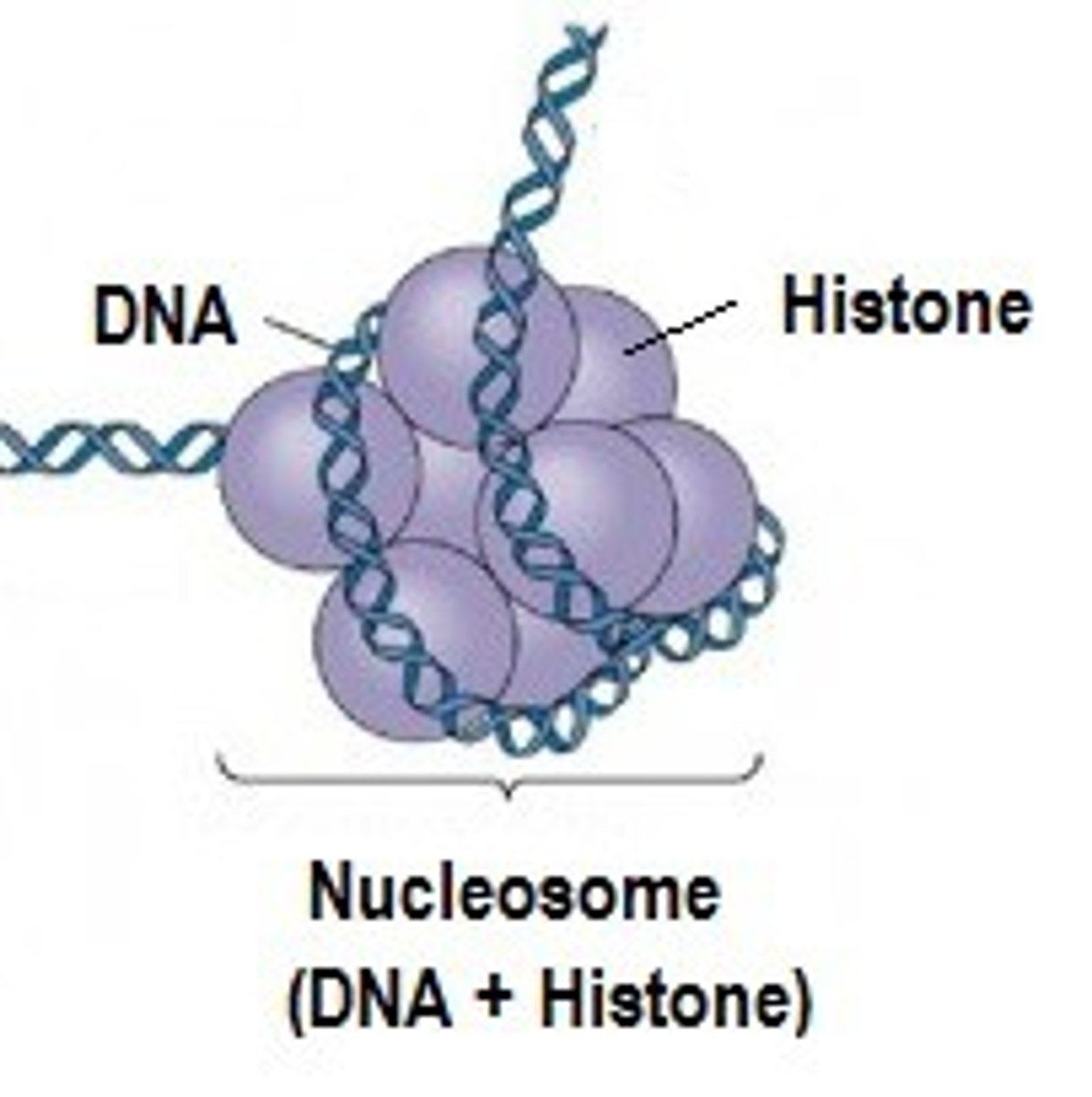
2 of each = 8 total proteins!
2 H2A, 2 H2B, 2 H3, 2 H4
How many core histones are in each nucleosome? How many total proteins make up the core histone?
Linker histone
Class of histone consisting of 1 protein. Consists of ~200 amino acids. Tissue specific expression and NOT highly conserved. Loosely associated with the histone core.
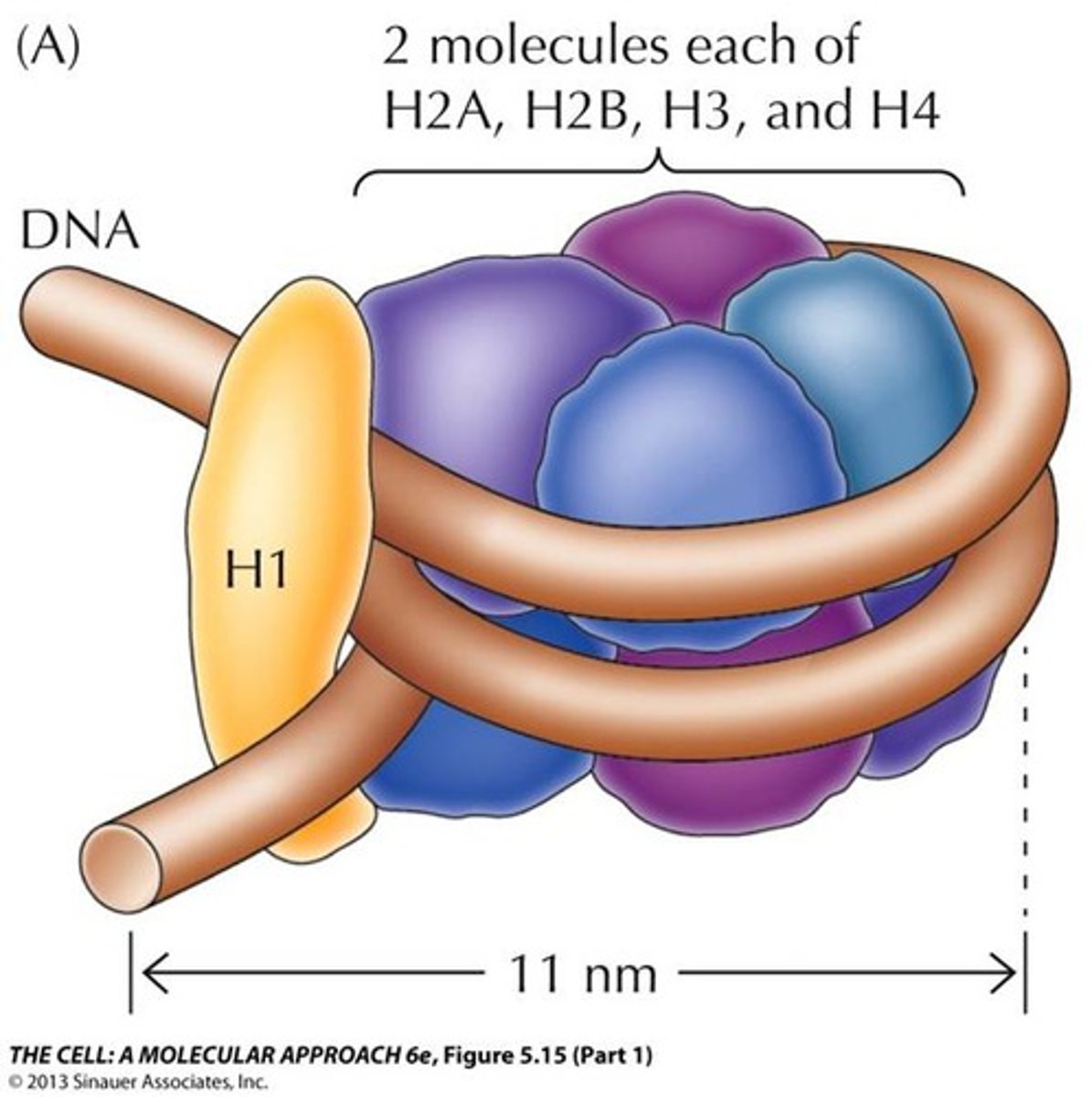
H1
What proteins form the Linker histone?
They have different sequences throughout the body
What is meant by "linker histones are NOT highly conserved"?
No, it is NOT part of the histone core, but it is part of the nucleosome
Is the linker histone part of the histone core?
146 base pairs
How many base pairs of DNA wrap around the histone core to form the nucleosome?
54 base pairs. Linker DNA
How many base pairs link one nucleosome to the next? What are these base pairs called?
Histone tails. They they can make chromatin more relaxed (euchromatin) or more condensed (heterochromatin), thus regulating the accessibility of DNA for transcription, replication, etc.
What part of the histone core, in relation to core histones helps determines the structure of chromatin? How?
7
Nucleosomes reduce DNA length by _____ times.
Linear. They look like beads on a string.
Describe the arrangement of nucleosomes.
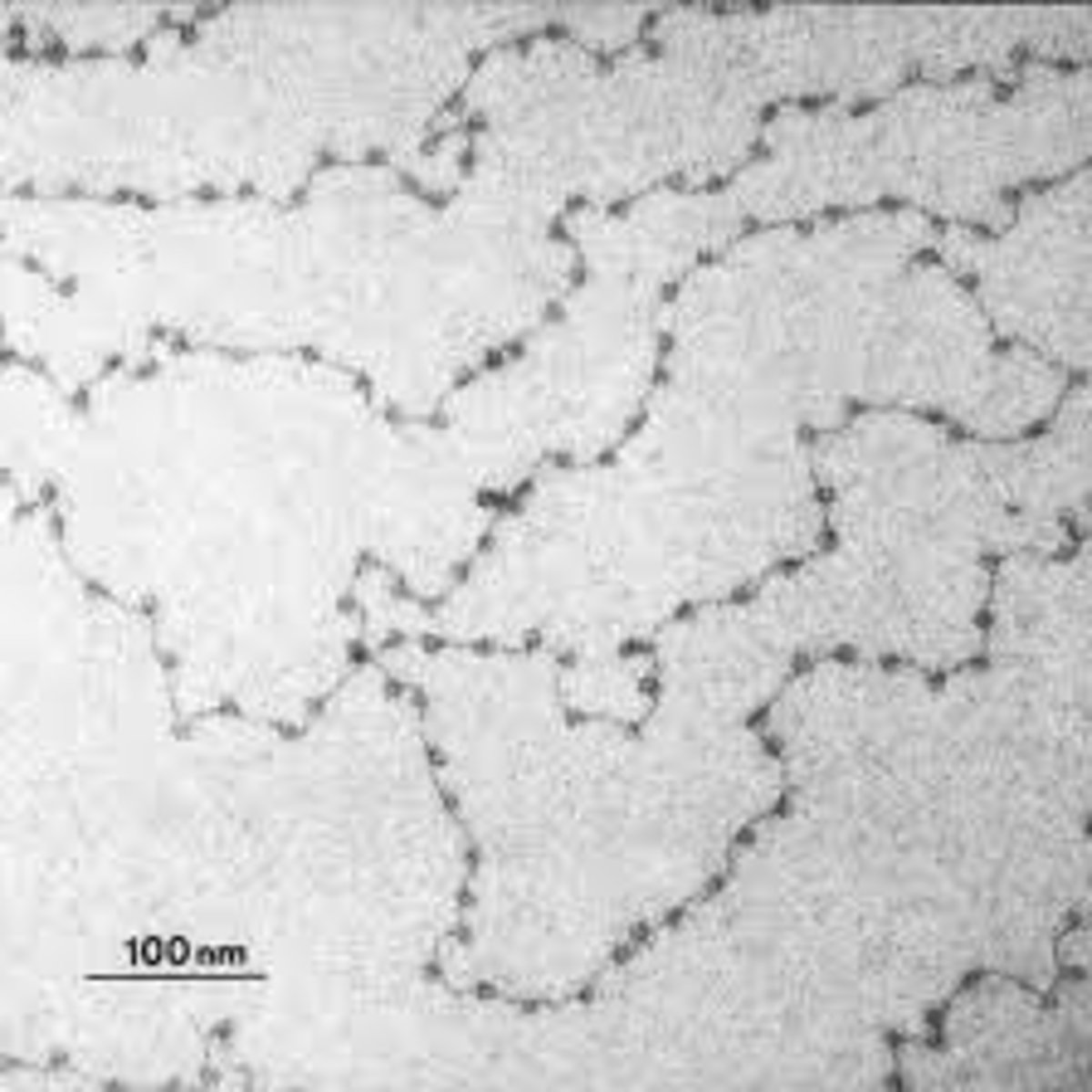
10 nm fiber
What is the approximate fiber diameter of each length of nucleosomes ("string of beads") that packs into the next chromatin structure?
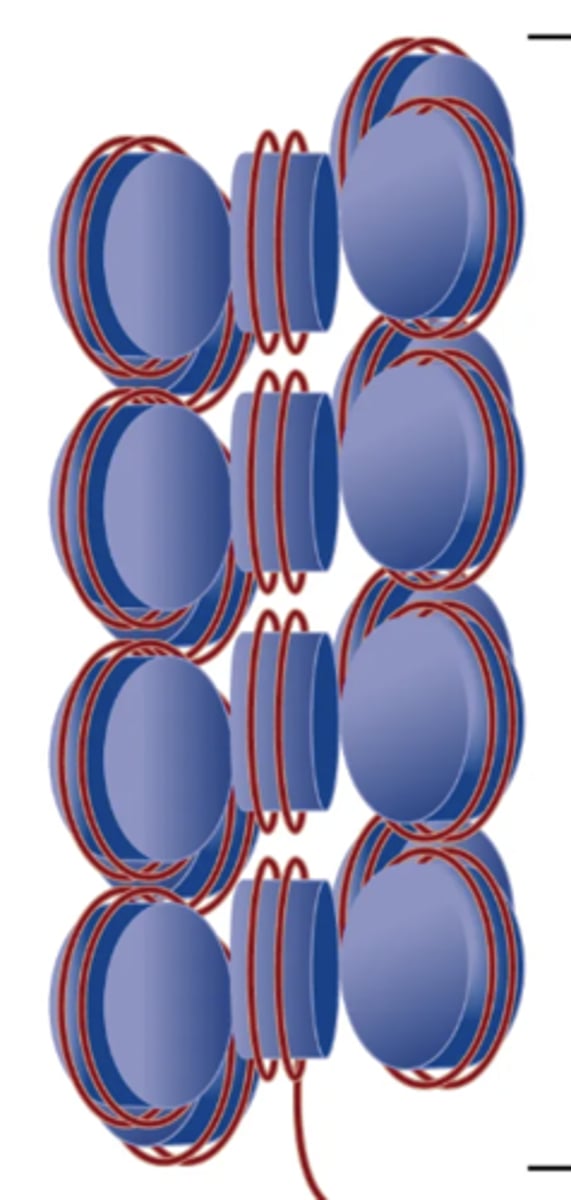
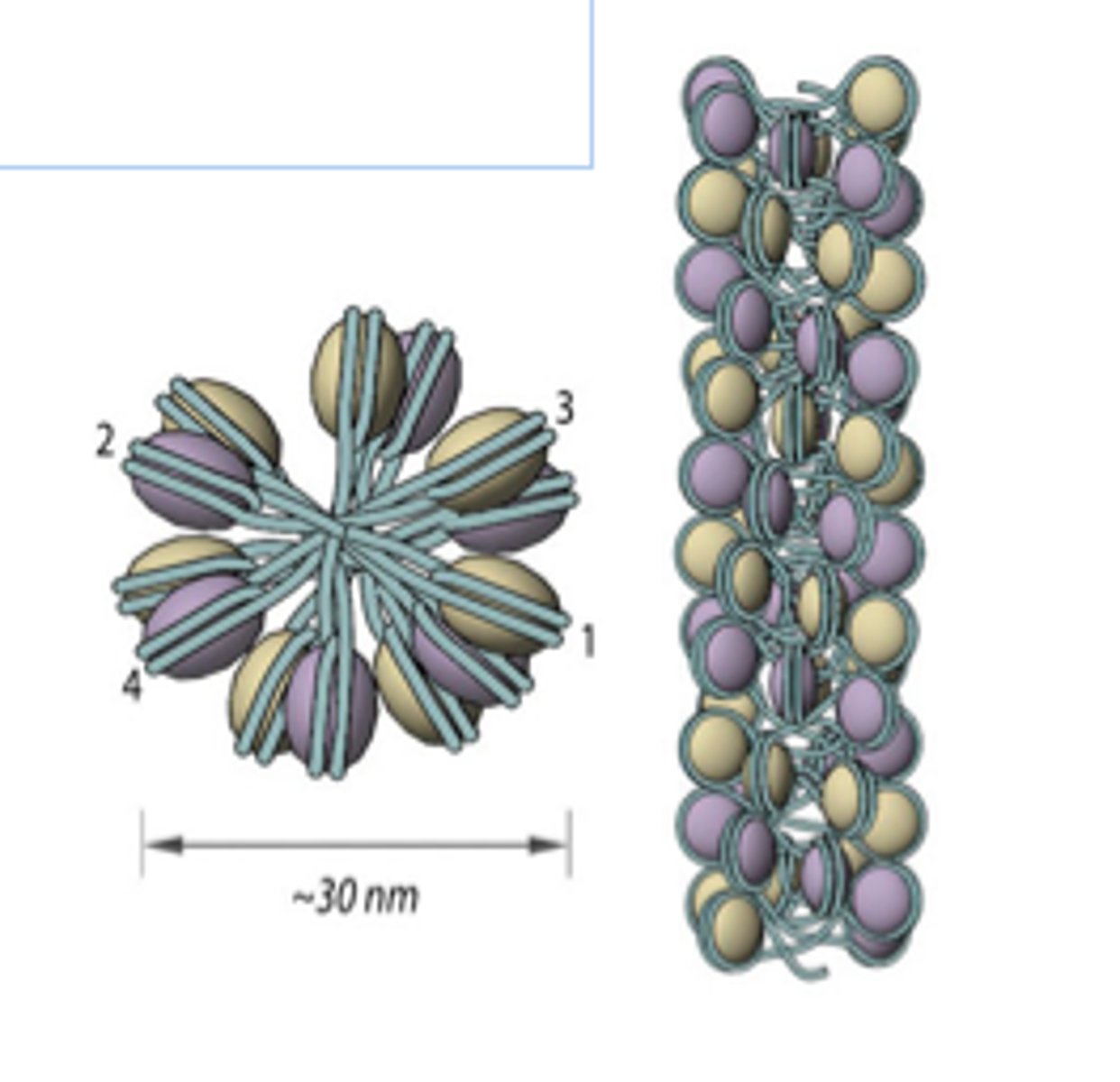
Solenoid Model
Supercoiling DNA model where a structure of multiple strings of nucleosomes packed together to form chromatine. Uses helical coiling of 10 nm fibers consisting of 6 nucleosomes stacked on top of each other in a helical arrangement to form one turn of this model. Nucleosomes have a uniform orientation. DNA has a 30 nm nucleosomal arrangement overall.
Considered the next step of chromosome formation after nucleosome formation.
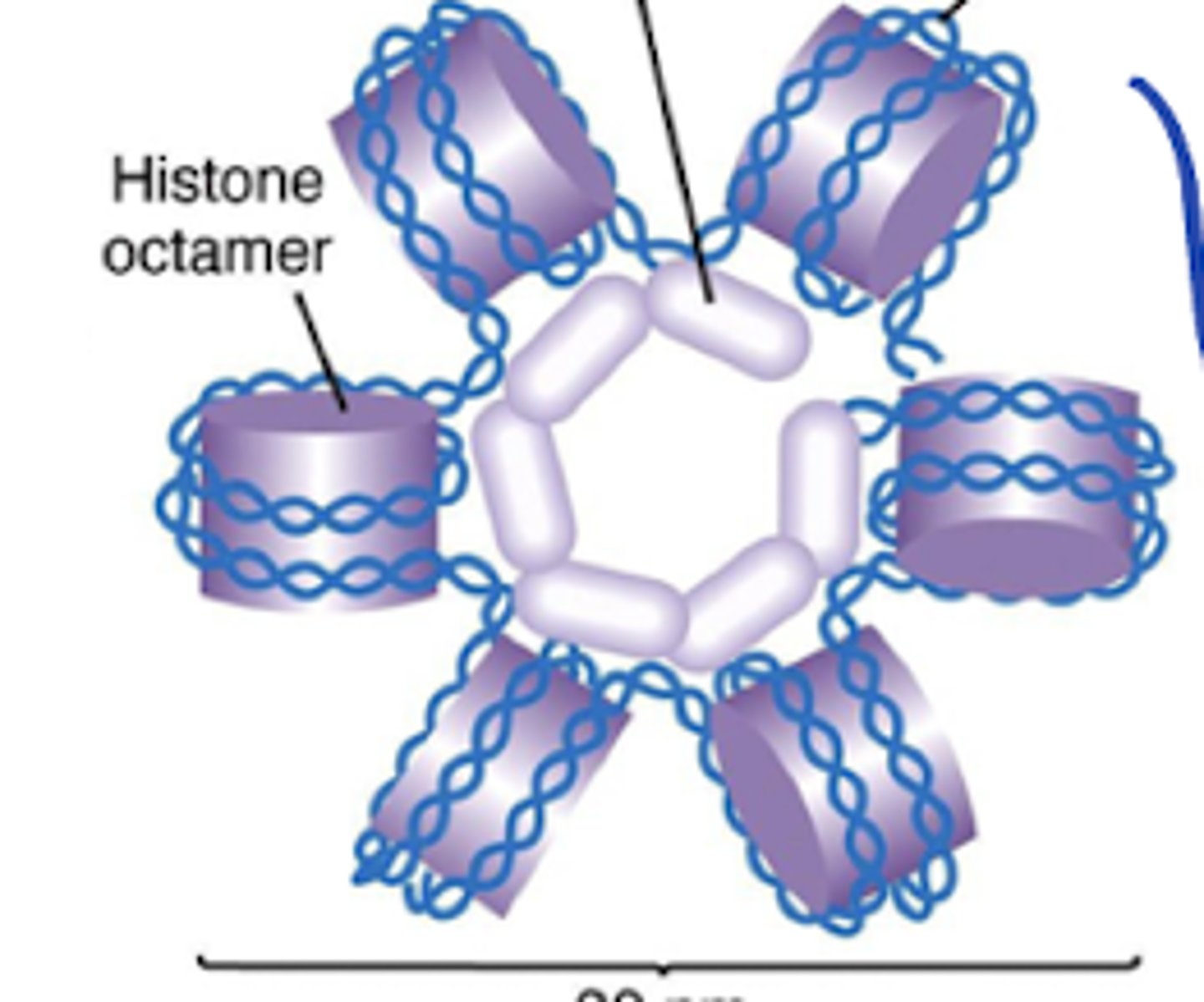
H1 histone/Linker histone
What structures of the nucleosome/which histone allows the nucleosomes to pack together in a ring-like formation to form the solenoid structure?
H1 binds to both linker DNA and the DNA wrapping around the histone (146 base pairs) and induces tighter DNA wrapping around histones to form the solenoid complex.
Compacts it by 40x.
How does the H1 histone/Linker histone work to bind nucleosomes together and form the solenoid? How much does it compact the DNA?
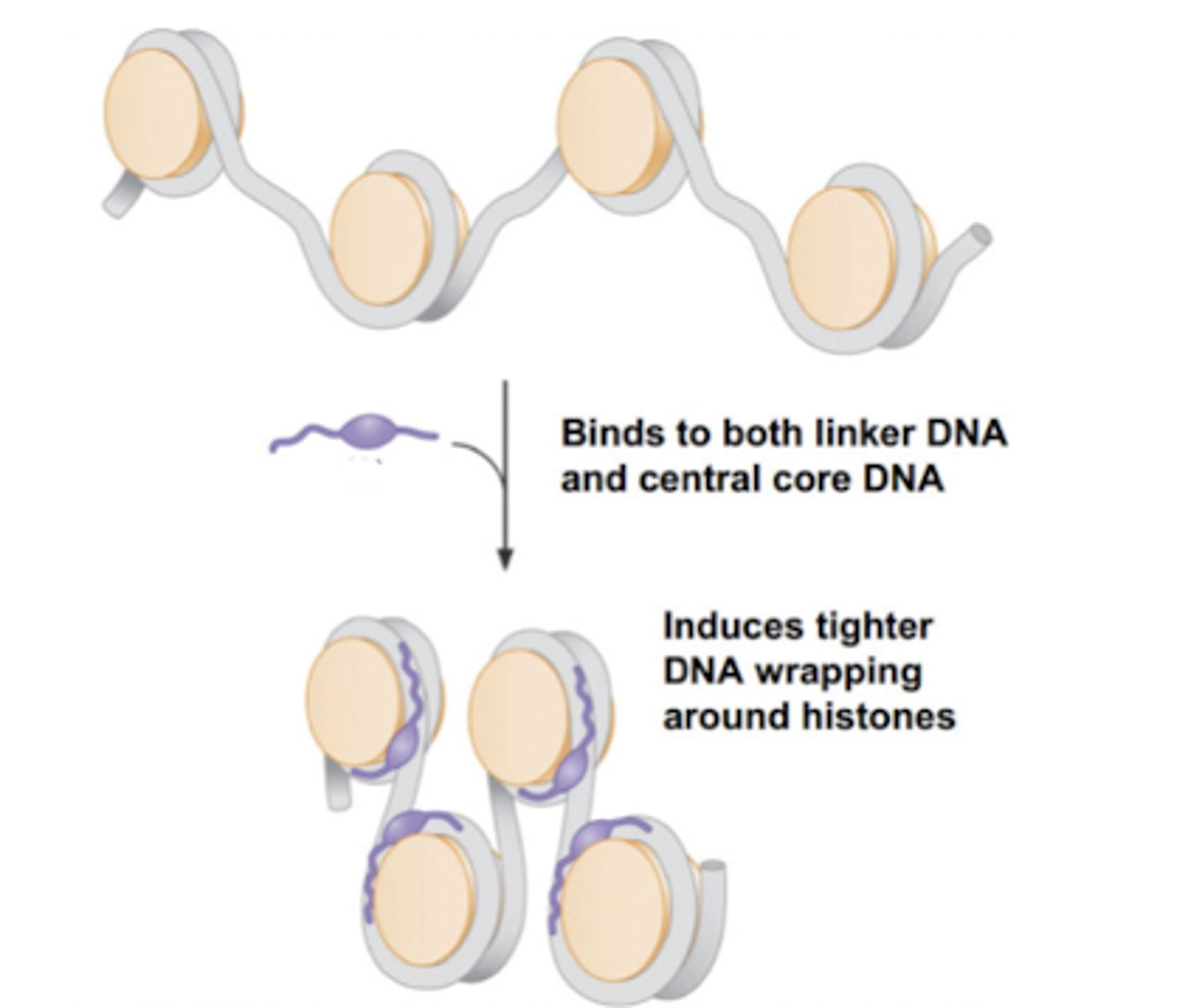
Histone H1
Adding ___________ induces compaction of DNA into the solenoid structure.
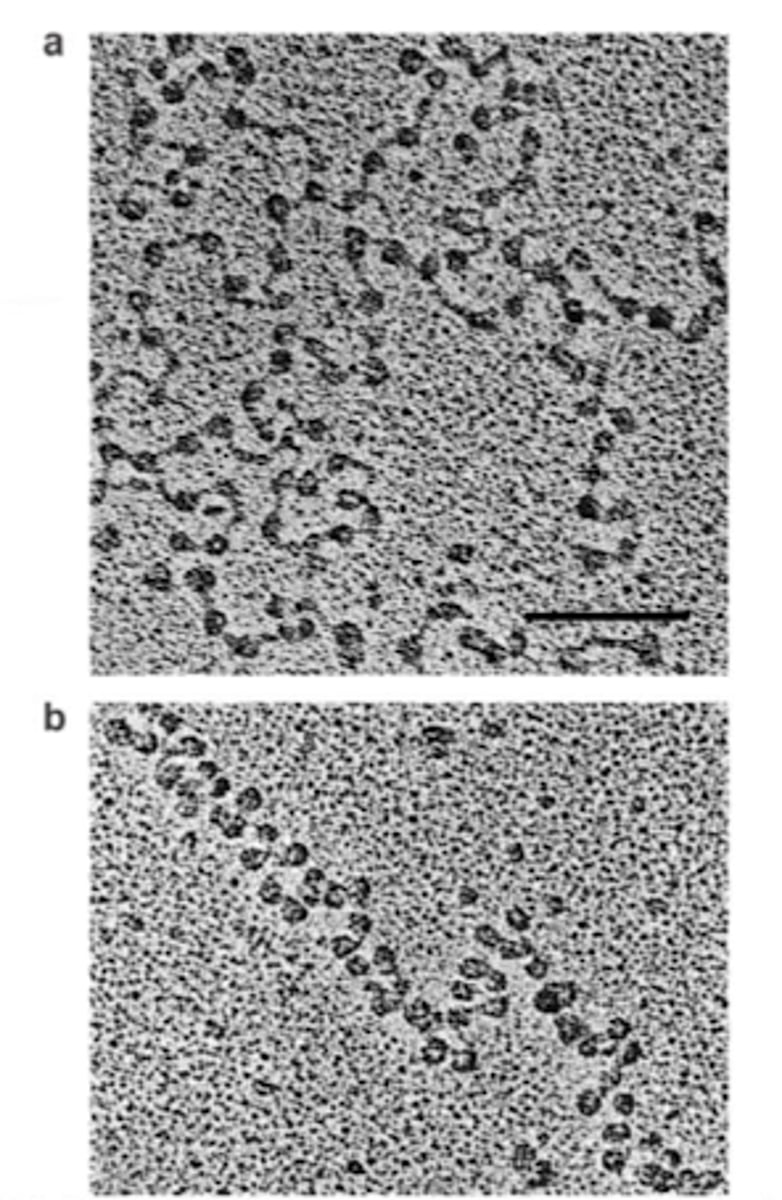
Zigzag model
The alternate supercoiling model of DNA that forms a chromatin structure using nucleosomes. Nucleosomes appear to be stacked in an nonuniform, zigzag pattern.
The DNA backbone is not flexible enough to bend between nucleosomes, so DNA CANNOT BEND in this structure. Allows more compaction than solenoid model.
Uses straight linker DNA to connect opposite nucleosomes.
True
T/F: experimental results show that both solenoid and zigzag topologies may both simultaneously be present in chromatin fiber
Chromatin loops
The next stage of chromosome formation after solenoid chromatin structures. DNA has a 300 nm fiber creating loops around a scaffold (center) made of topoisomerase II. Euchromatin is most present and compacted here.
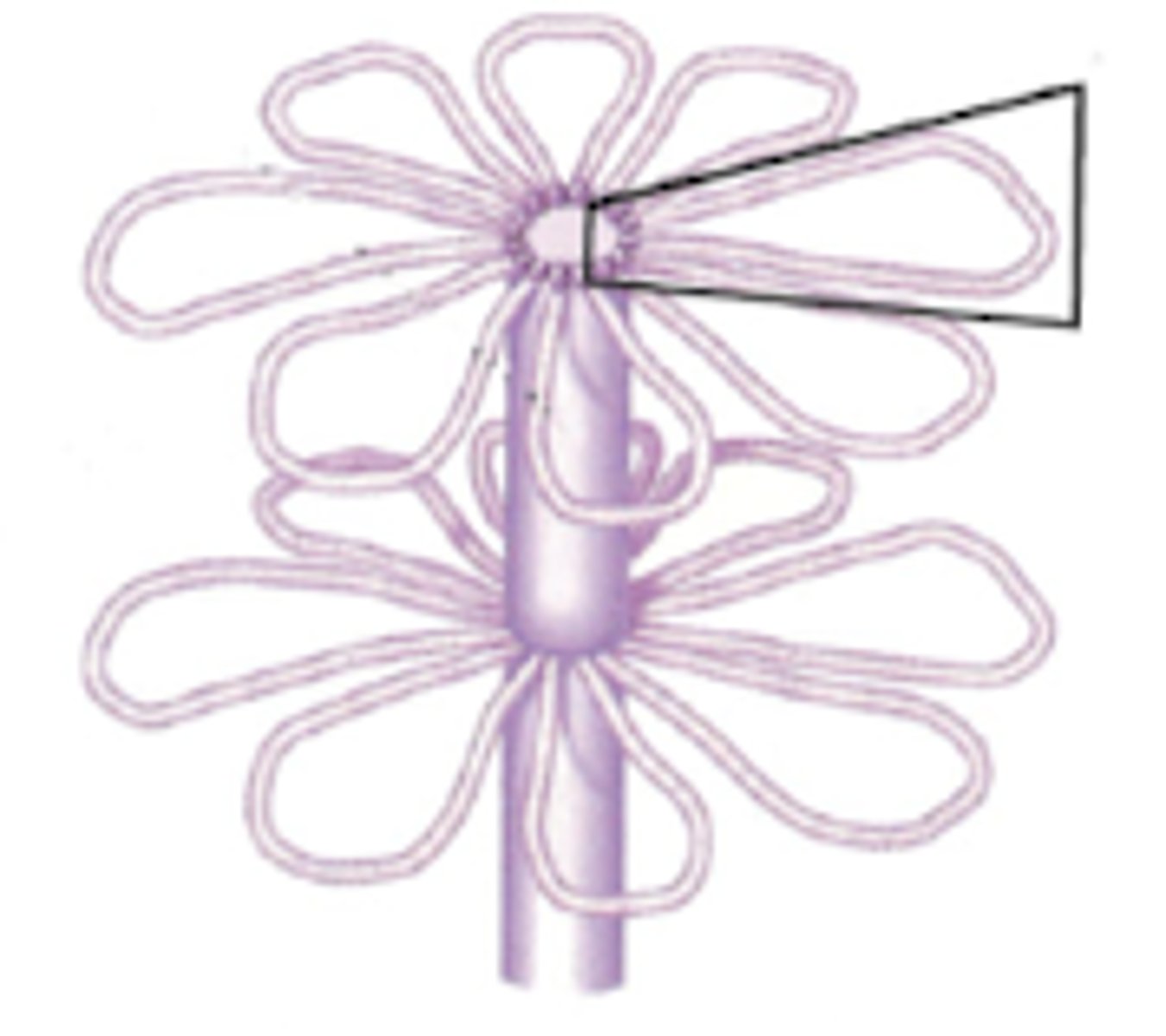
Condensed chromatin loops
Last stage of chromosome formation. Has a metaphase chromosome structure. DNA is 700 nm. Spiral scaffold composed of topoisomerase II and 15 non-histone proteins. Heterochromatin is most present and compact here.

1. G1 Phase (cell grows)
2. S Phase (cell replicates)
3. G2 Phase (cell grows prepares for mitosis)
4. Mitosis
List the different phases of the cell cycle.
1. G1 Phase:
2 nm-10 nm DNA fiber (DNA strands and nucleosome formation)
2. S Phase:
No formation here
3. G2 Phase:
30 nm fiber (Solenoid chromatin structure)
300 nm fiber 700 nm fiber (Chromatin loops)
700 nm fiber (very end of G2) (Condensed chromatin loops)
4. Mitosis
Whole chromsome
List the diameters of DNA in each phase of the cell cycle.
Karyotype
A display of the chromosome pairs of a cell. Arranged by chromosome size, centromere position, and whether the chromatin is heterochromatin vs euchromatin.
Complexity
the intricate ways in which genetic information is organized, regulated, and expressed, leading to the diverse traits and functions seen in organisms.
The number and type of cells and the degree of cellular organization.
What defines complexity within organisms?
hierarchial
Organisms have a _________ building of complexity. They build upon each other from atoms to cells to living things.
in eukaryotes, just as the number of genes does not correlate well with organismal complexity, the size of the genome is unrelated to the metabolic, developmental, and behavioral complexity of an organism
SO, they are NOT correlated
Does the amount of DNA in a genome correlate with the complexity of the organism?
Bc of "junk DNA". A majority of organisms with a large amount of DNA use very little of it, while the majority of it goes to waste.
Ex. flatworms
WHY does genome size alone not correlate with complexity?
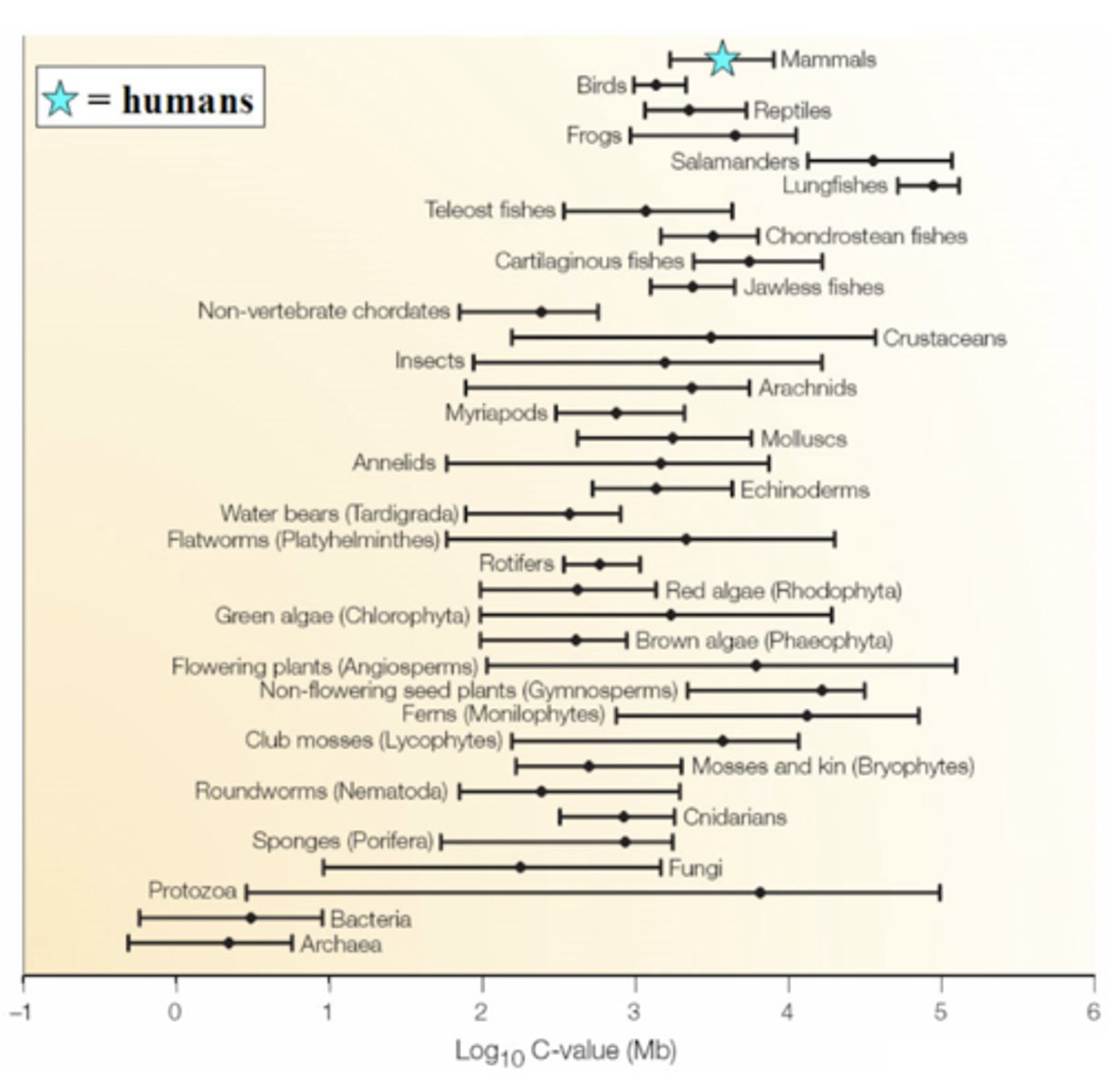
C-value paradox
States that genome size (INCLUDES "junk DNA" AND coding DNA) does not correlate with organismal complexity.
ex. frogs may have more DNA than humans, but that does not make them more complex than humans.
the Onion Test
Test that showed that onion has 5x more DNA than humans, but that does not make it more complex than humans.
G-value Paradox
The number of protein-coding genes ALONE does not correlate with organismal complexity.
Does NOT include junk DNA
ex. humans have around 20,000-25,000 protein-coding genes, but so do nematodes and watercress
While onions may have more DNA, than humans, we can do more with our DNA than onions (ex. have the ability to splice our genes, can make more proteins with fewer material), so we are more advanced
Why are humans actually considered to be more advanced than an onion?
1. Highly repetitive DNA sequence (HR)
2. Moderately repetitive DNA sequence (MR)
3. Single Copy DNA sequence (Unique)
List the 3 types of DNA in the genome
Highly repetitive DNA sequence (HR)
Type of DNA that comprises 10% of human genome.
Found in heterochromatin/"non-coding" DNA regions.
Present at variable lengths of base pairs (5-100 bp).
Made of > or equal to 10^6 copies per genome.
ex. alpha satellite DNA
Alpha satellite DNA
Type of repetitive DNA with a high order of repeated base pair sequences.
Each repeated base pair monomer is 171 base pairs long (can be any repeated arrangement of base pairs). When these repeated base pair monomers come together to form one sequence, they become a higher order repeat (HOR).
A HOR might consist of a specific arrangement of 10-20 monomers repeated hundreds or thousands of times.
When the HOR's come together, they form an HOR alpha satellite array consisting of multiple HOR sequences.
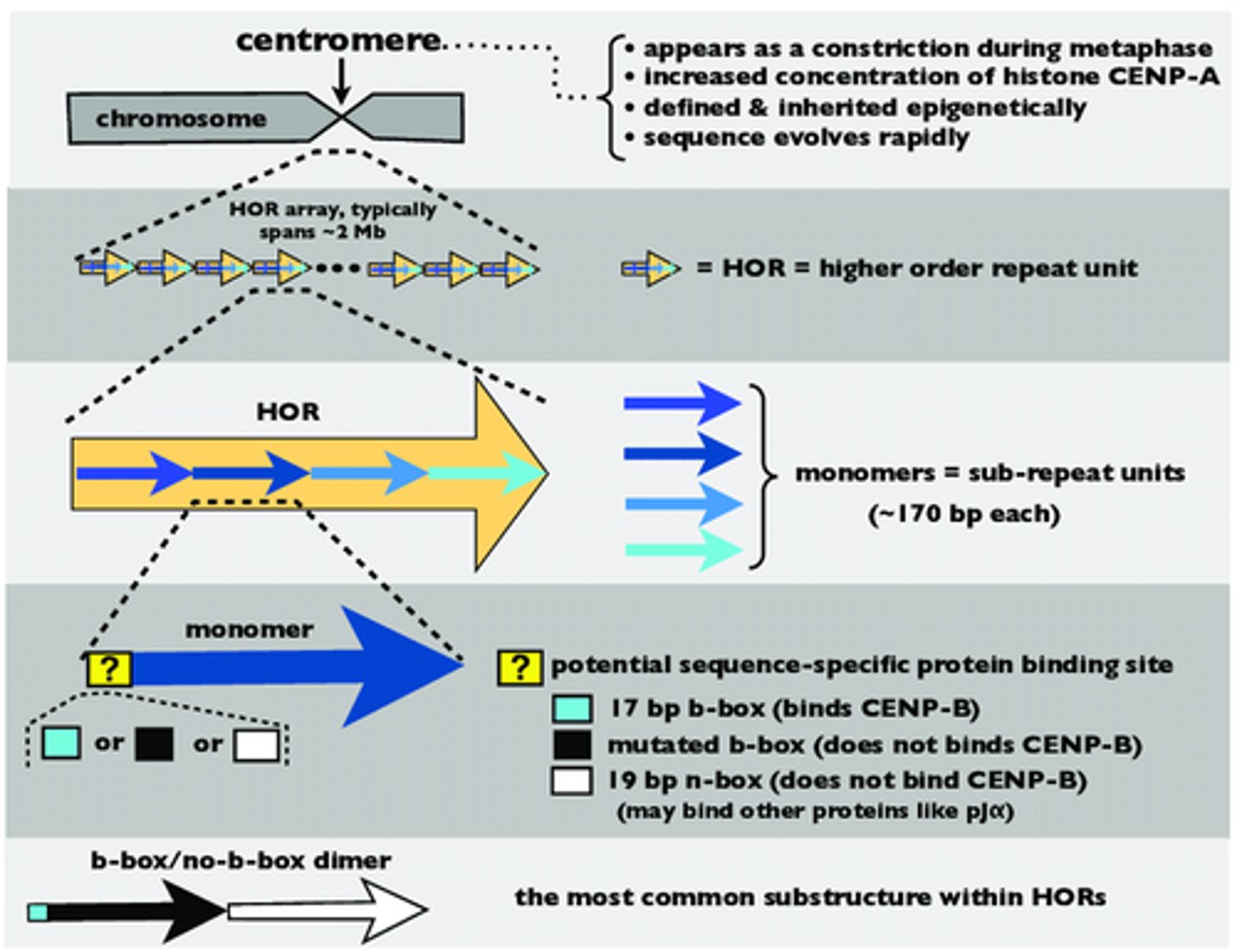
monomeric
Anything not in the repeated base pair sequence of alpha satellite DNA is _________
Ranges from structural and organizational roles to junk DNA
What is the range of functions for HR DNA?
true
T/F: HR DNA is mostly found around telomeres and centromeres, but can be found anywhere in the chromosome
Moderately repetitive DNA (MR)
Type of DNA sequence that comprises 30% of the human genome. Scattered throughout euchromatin/"coding" areas.
Average 300 bp in size.
Present between 10-10^5 copies per genome.
Includes "redundant" genes
ex. microsatellites
Redundant genes
Genes that you need in MASS quantities to operate the cell, ex. histones, RNA, proteins.
Microsatellites
Type of DNA strand with short repeated sequences. Has useful genetic markers as they tend to be highly polymorphic.
1. di
2. tri
3. tetra
List the 3 types of microsatellites.
Di-microsatellite
Microsatellite with 2 base pairs per repeat sequence

Tri-microsatellite
Microsatellite with 3 base pairs per repeat sequence

Tetra-satellite
Microsatellite with 4 base pairs per repeat sequence

Slippage recognition
Microsatellites occur through a mutation known as?
1. used to sequence human genomes
2. used as markers for certain disease conditions
3. primary markers for DNA testing in forensic cases
List 3 functions of microsatellites in research.
Single Copy DNA sequence
AKA unique DNA. Compromises 1-5% of the human genome. Found in euchromatin/"coding" areas.
Codes for proteins that are not always needed - we turn them on when needed and off when not.
Contains 20,000 protein-coding genes.
Present at a single or low copy number per gene.
It is unknown
If HR, MR, and Unique DNA add up to form 45% of the genome, what is the other 55% made from?
Gene
The basic and functional unit of heredity. Sequence of unique nucleotides (genotype) that carry the genetic information which is to be expressed (phenotype).
"Instruction manuals for our bodies"
2
Every person has ____ copies of each gene - one from mom, one from dad
> 99%, < 1%
> ______% of the number of genes are the same in ALL people. < _____% of the total number of genes contribute to each person's unique physical features
Molecular level gene
DNA sequence that gives rise to an RNA molecule
transcriptional unit
What we see when looking at gene on a molecular level. A segment of DNA that is transcribed into RNA as a single molecule. It encompasses all the necessary elements required for the transcription process, including promoters, terminators, coding sequences, and regulatory sequences.
1. exons
2. introns
What are the two coding areas of the transcribed region of RNA (not mRNA)?
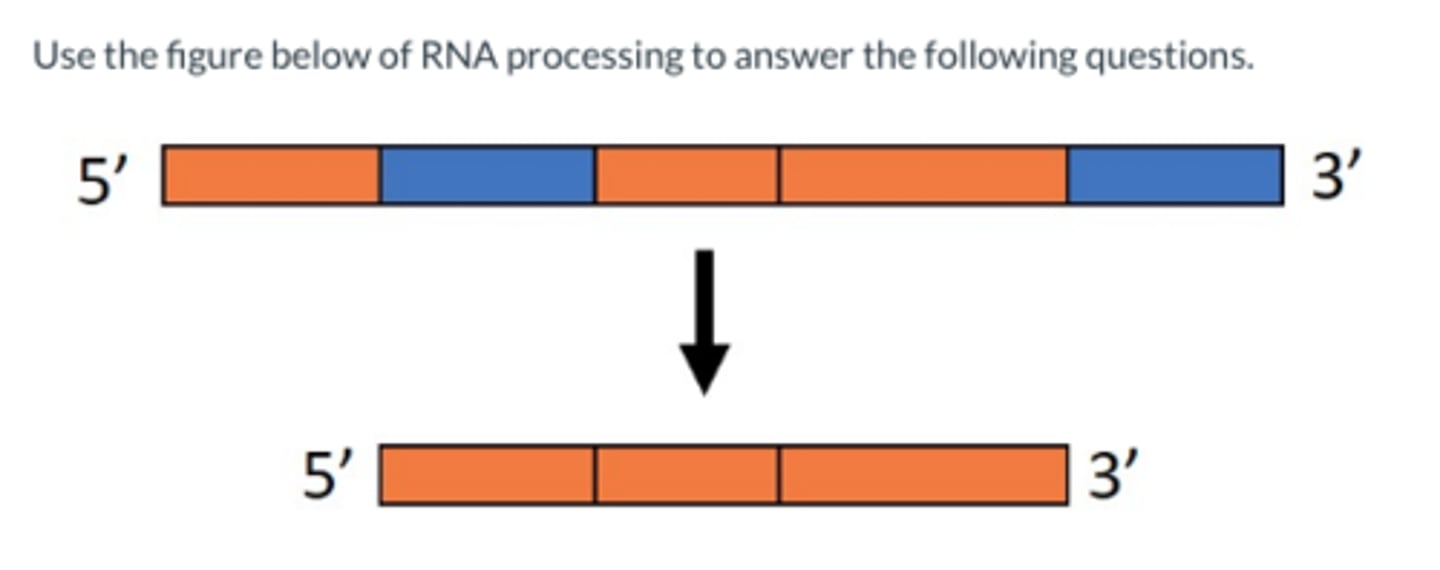
Exon
"Coding sequence" of RNA = phenotype. They are both transcribed and translated.
Intron
"Intervening sequence" of RNA = do NOT code for phenotypes. They are transcribed from DNA, but not translated because they are removed from RNA --> mRNA
1. 5' untranslated region
2. 3' untranslated region
List the flanking regions of the transcribed region of RNA
5' untranslated region
Flanking region where mRNA is directly upstream from the initiation codon, so it is not translated to mRNA
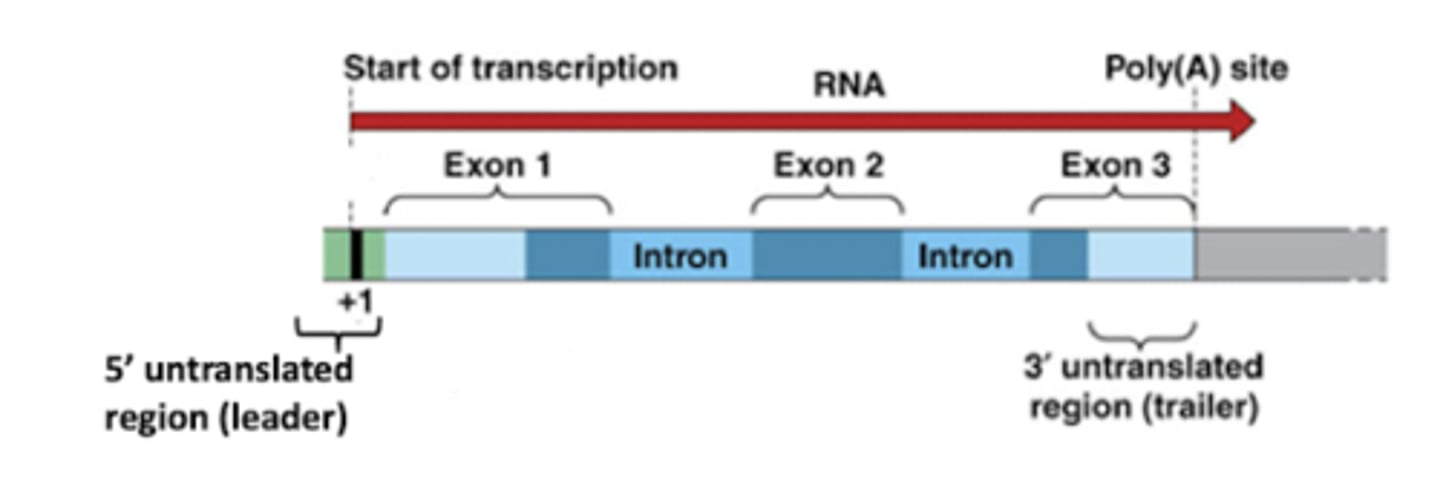
3' untranslated region
Flanking region where mRNA immediately follows the translation termination codon
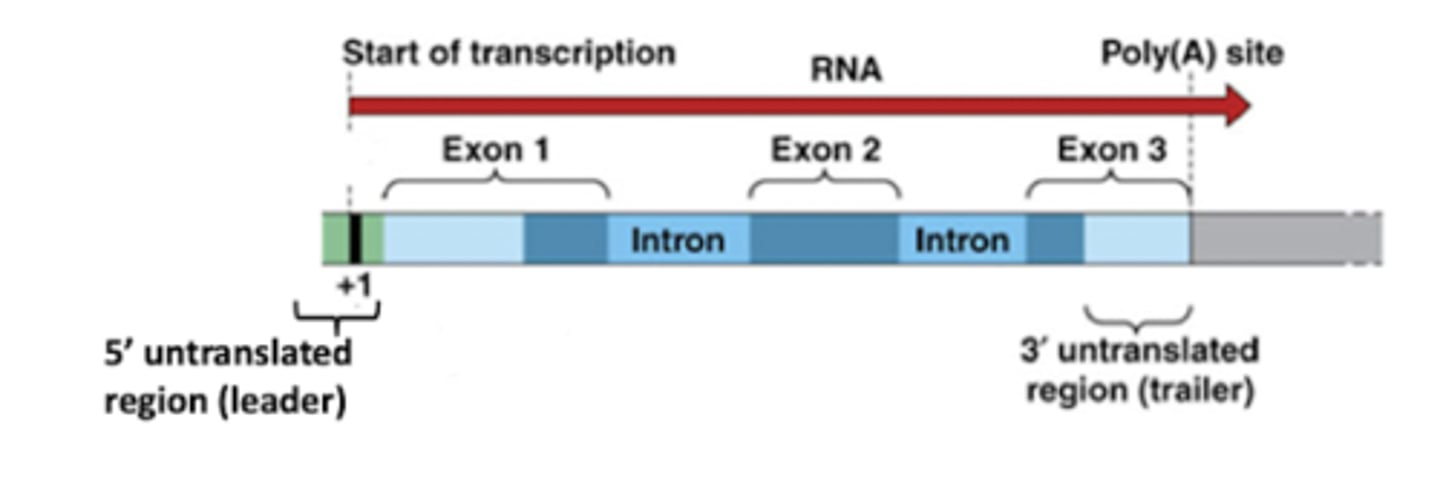
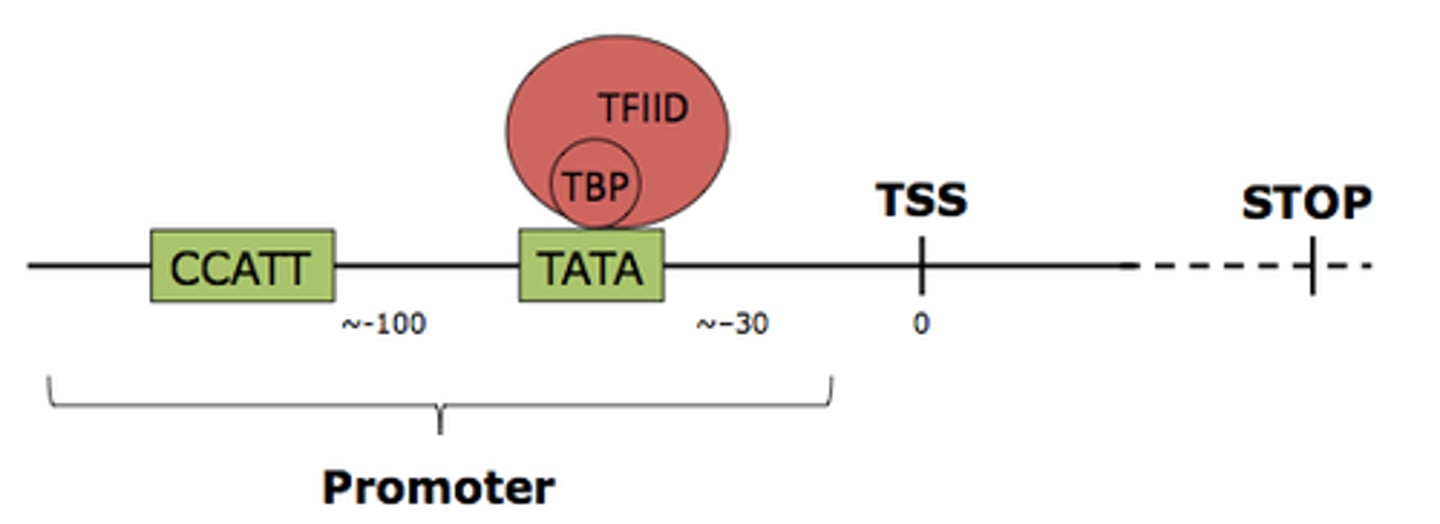
Promoter
DNA sequence onto which transcription machinery binds and initiates transcription. Includes TATA box.
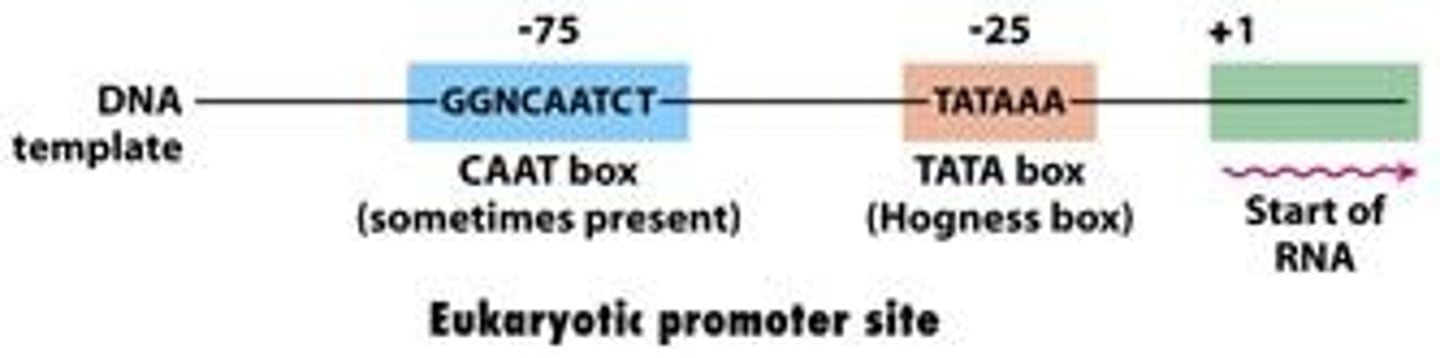
TATA Box
Highly conserved DNA sequence of TATA base pairs that serves as the binding site for transcription factors to turn translation on/off. Located about 25-30 bases upstream of transcription site. Part of the promoter region.
Enhancer
A segment of eukaryotic DNA containing multiple control elements. They function by binding specific proteins, such as transcription factors, to increase the transcription of a target gene, often located at a significant distance from the enhancer itself.
1. CAAT box
2. CG box
List two types of enhancers primarily found on DNA within eukaryotes.
CAAT box
Type of enhancer that occurs 60-100 bases upstream to the transcription site.
Has a consensus sequence of 5'-GGCCAATCT-3'. The core sequence, CAAT, gives this element its name.
Typically required for inducible genes to be produced in large amounts.
CG box
Type of enhancer that occurs ~100 bases upstream to the transcription site.
Region of DNA that can be bound with proteins (activators) to activate transcription of a gene or genes
Terminator
Section of nucleic acid sequemce that marks the end of s gene or operon in genomic DNA during transcription. Occurs AFTER the poly-A tail.
1. Solitary genes (unique)
2. Duplicated genes
3. Multigene Families
4. Pseudogenes
5. Repeated Genes
List the 5 different types of genes.
Solitary genes
Type of gene where there is a single copy in a haploid genome and two copies in a diploid genome. Comprises the bulk of euchromatin
Duplicated genes
Type of gene made by a portion of a chromosome being duplicated and resulting in an additional copy of the gene. This copy is called a paralog gene.
Either of the two genes may mutate and change the original function of the gene. Usually occurs due to an error in meiosis.
Multigene Families
A set of several similar genes, formed by duplication of a single original gene, and generally within similar biochemical functions. Most often located in similar areas of the chromosome. Most often used or synthesized at different times.
Pseudogenes
Dysfunctional relatives of genes that have lost their protein-coding ability. Often the result of multiple mutations within a gene
Repeated genes
Multiple copies of small genes clustered through the genome at specific sites. Present in a high copy number. Many times present in a "head to tail" conformation.
repeated
Genes for tRNA and rRNA are __________ genes.
Central dogma
DNA -> RNA -> Protein.
1 Gene (genotype) codes for 1 mRNA (intermediate), 1 mRNA codes for 1 protein (phenotype)
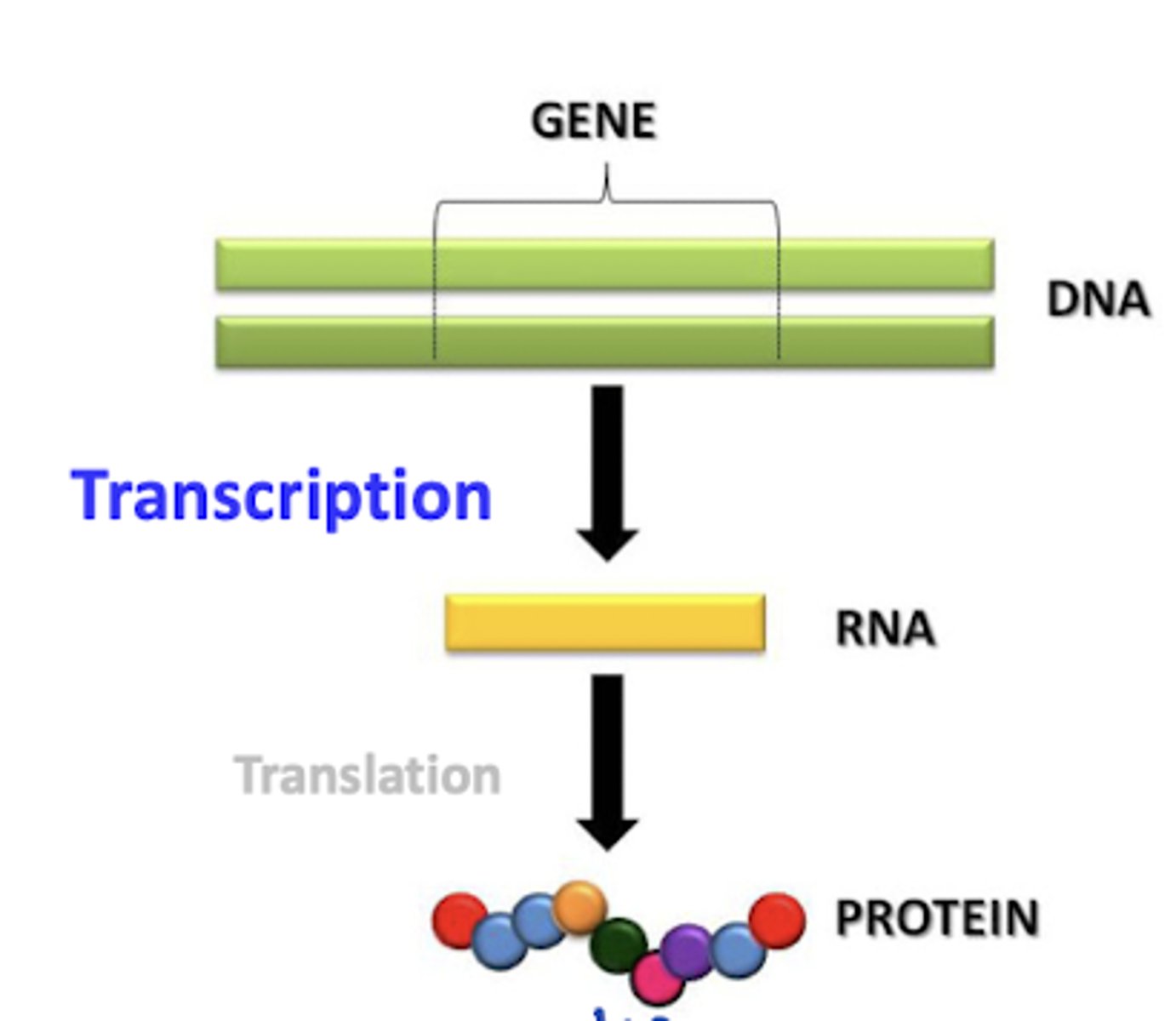
Transcription
DNA --> mRNA process.
Occurs in the nucleus of eukaryotes and in the cell of prokaryotes.
Occurs during either G1/S or G2/M.