unit 5: mitochondria, ETC, mtDNA
1/76
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
77 Terms
how does the mitochondria of eukaryotes produce ATP?
burning food by oxidative phosphorylation
what makes oxidative phosphorylation an efficient means of ATP generation?
the sufficient amount of oxygen gas
what is the only amitochondriate eukaryote known?
Monocercomonoides
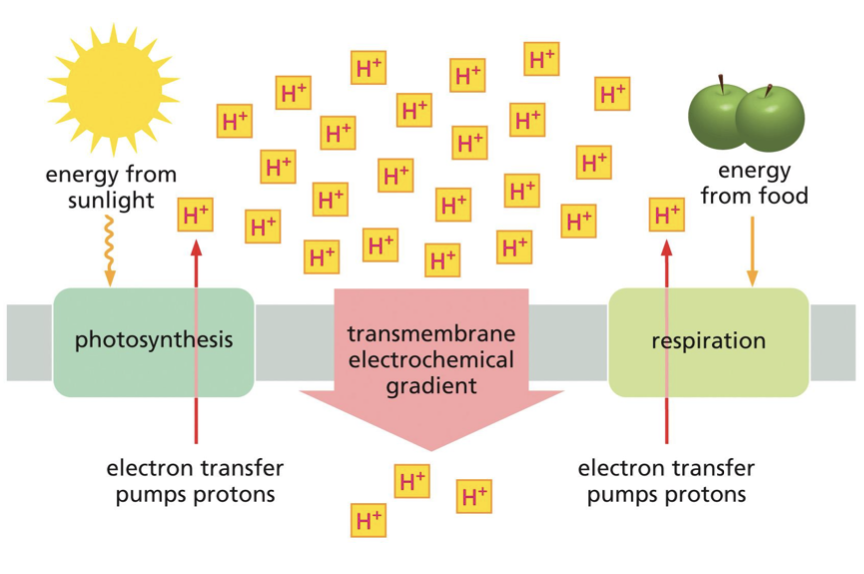
what is the shared mechanism for harnessing energy between chloroplasts and mitochondria?
chemiosmotic coupling
define and describe chemiosmotic coupling
high energy electrons from oxidation and photon excitation are transferred through a series of membrane-embedded protein complexes that form the electron transport chain (ETC). each electron transfer releases a small amount of energy via hydrogen ions being pumped across the membrane: this generates an electrochemical gradient. protons flow back down the electrochemical gradient through the ATP synthase, which converts ADP and P into ATP.
is the mitochondria static in structure?
no, it is constantly changing shape via division and fusion. its structure depends on the cell type
what dye is used to observe mitochondria?
mitotracker red, which is a fluorescent dye taken up into mitochondria on basis of membrane potential across the inner membrane
live cell imaging can also be done using rhodamine 123, which is a dye specific for mitochondria. rhodamine 123 is following by fixation and staining with fluorescent antibodies against microtubules to allow for overlap visualization (picture)
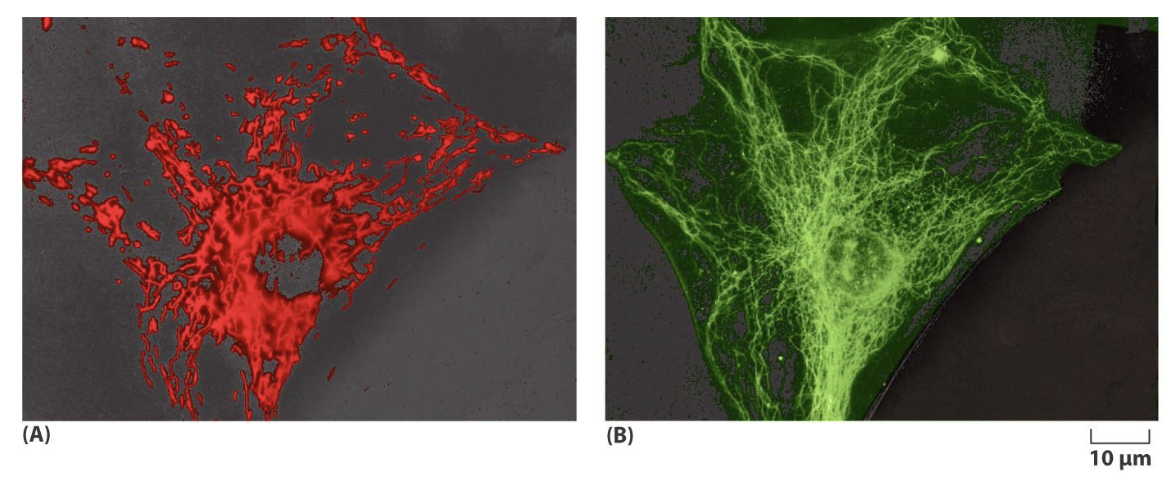
what contributes to distribution and orientation of mitochondria in a cell?
its association with microtubules
where would the mitochondria be found in cardiac and skeletal muscles?
between myofibrils because it has a high demand for ATP
where would the mitochondria be found in sperm cells?
wrapped around flagellum due to high demand for ATP
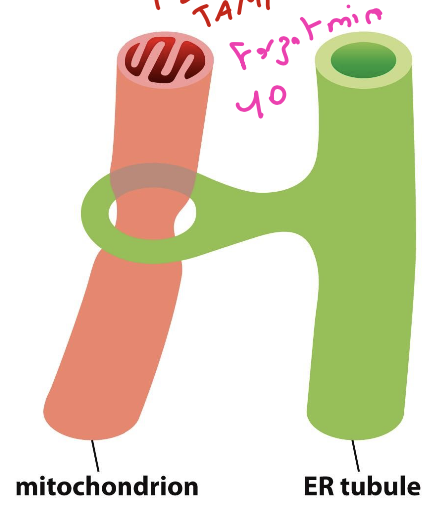
define mitochondria-associated membrane (MAM) and how you could view it
a physical connection between the mitochondria and ER tubules that determine site of division for mitochondria.
how to view: tag the ER with an anti-Sec61 antibody and use anti-TOM/TIM antibody with another fluorescence to view co-localization of the two
how many membranes are mitochondria bound by?
2. they’re the only organelle bound by 2, which allows them to have 5 functional spaces
list the 5 mitochondrial spaces in order from outermost to innermost with a brief description of what goes on in them
outer membrane (OMM): has lots of porin, TOMs, and pro-apoptotic proteins
intermembrane space (IMS): is equivalent to the cytosol in terms of pH and ion composition. it has no electrochemical gradient
inner membrane (IMM): machinery for oxidative phos, metabolite transporters, TIMs all found here. this area generates cristal space
matrix: has metabolic enzymes and performs oxidation of foodstuffs
cristal space: traps protons
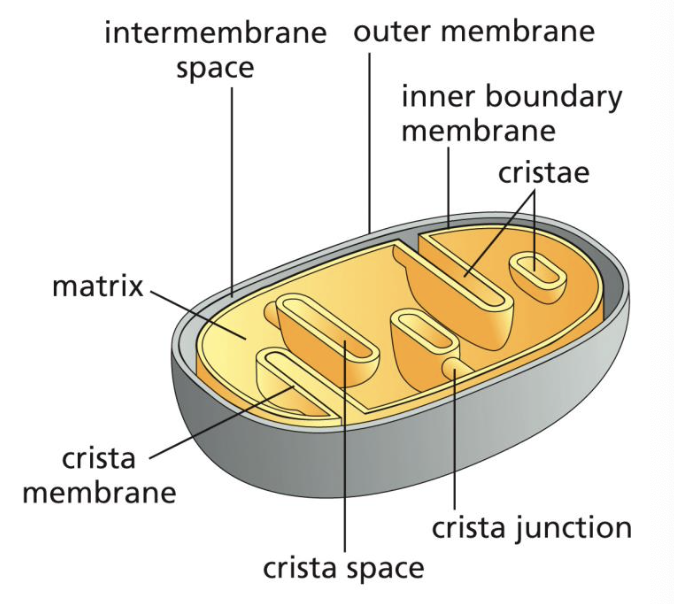
what are cristae?
membrane discs that protrude deeply into the matrix with an interior consisting of the cristal space. the crista membrane is contiguous with the inner mitochondrial membrane and contains the respiratory chain complexes and ATP synthase.
what is the inner boundary membrane?
the membrane where the IMM runs parallel to the OMM
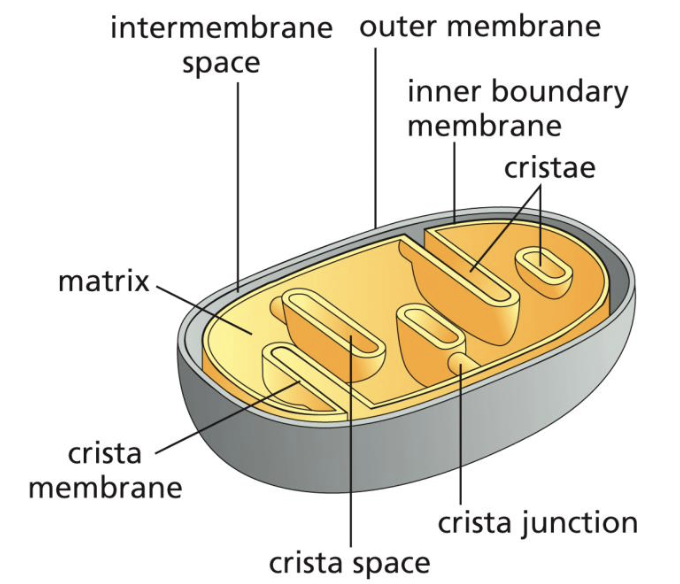
what mode of exchange does the IMM have?
diffusion of ions and small molecules
detail the fractionation process of mitochondria
in a low osmotic environment, an influx of water causes the mitochondrion to swell and the OMM will rupture, releasing the IMS contents
centrifugation of this product leaves the contents of the IMS in the pellet. the supernatant (OMM fragments + mitoplasts) is transferred to a medium of high osmolarity to cause shrinkage and form mitoplasts (a mitochondria that only consists of IMM and matrix)
density-gradient centrifugation separates the outer membrane from the dense matrix and its surrounding inner membrane. disruption and centrifugation separate the IMM from the matrix components
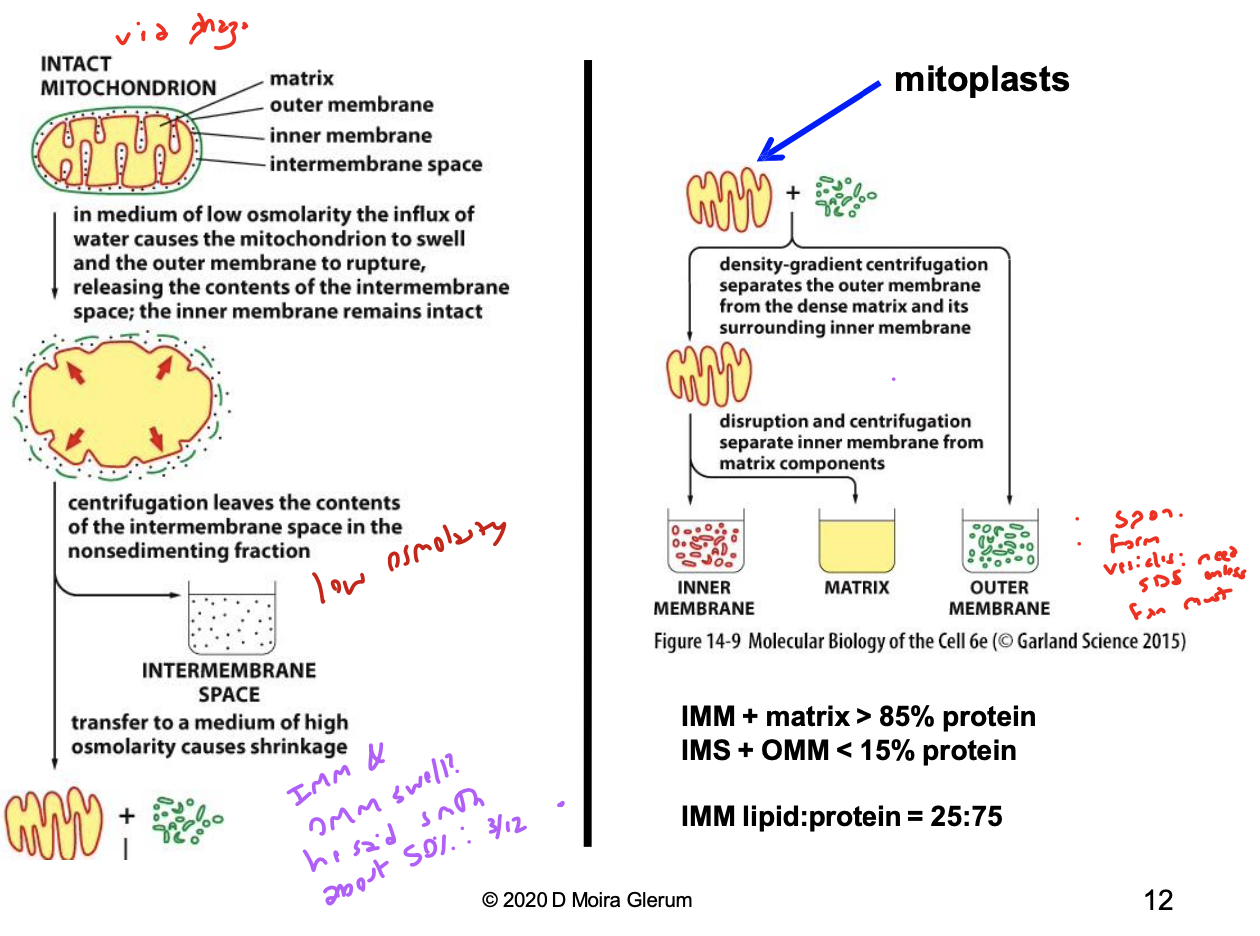
what %age of protein is found in the IMM and matrix?
85%
what %age of protein is found in the IMS and OMM?
15%
what is the IMM lipid to protein ratio?
25:75
what happens to the OMM sample after full fractionation of a mitochondrion?
it spontaneously forms vesicles, which means you’ll need SDS to get to the proteins in them
what does the mitochondria oxidize to produce energy? give a specific molecule and explain why it’s used
it oxidizes “foodstuff”, namely FADH2 because it donates very high energy electrons that feed into the citric acid cycle
you don’t need to know this cycle but know what’s involved
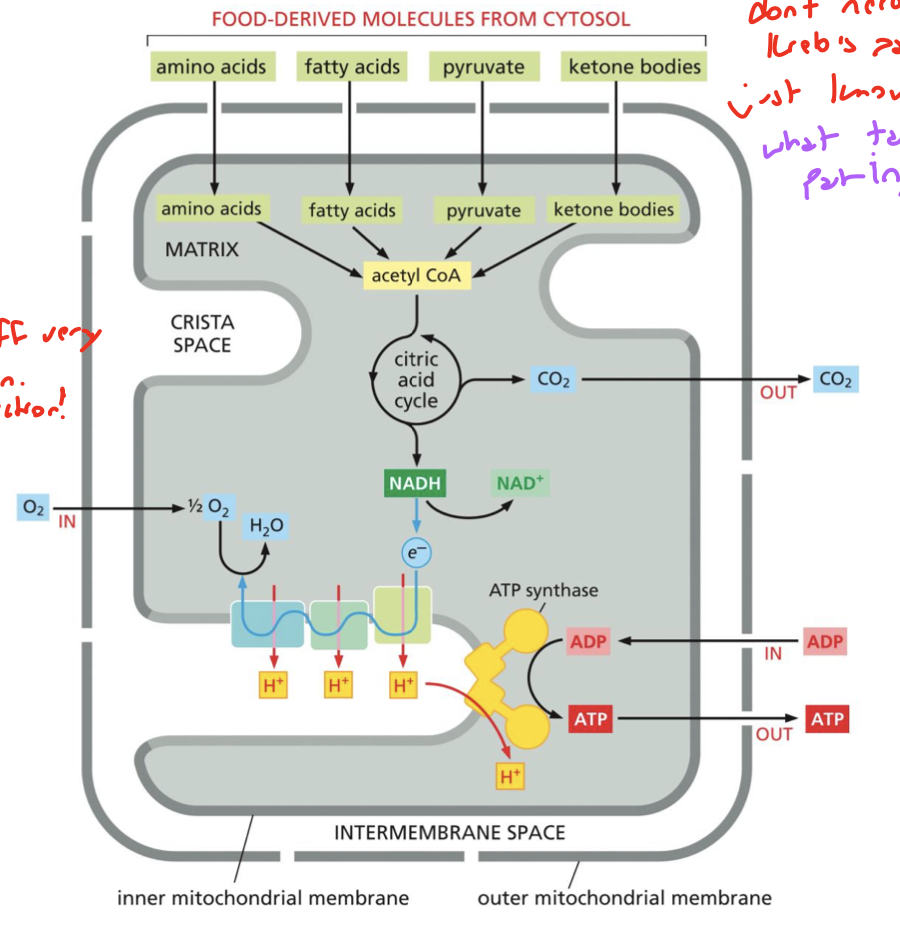
page 14 of unit 5.1 and min 60 of 3/12 lecture
aside from being a powerhouse, what does the mitochondria do?
provide the required carbon skeletons and reducing power to the cytosol
transports citrate to the cytosol to generate carbon and NADPH
buffers redox potential in cytosol so we can get NAD+ for glycolysis. this is done through metabolite shuttle systems since the IMM is impermeable to NADH
site of FeS cluster (the proteins involved in redox reactions), heme biogenesis, part of the urea cycle, lipid biosynthesis
plays an important role in calcium buffering
what does the mitochondria do during starvation conditions?
uses amino acids to fuel ATP prpduction
what do cells relying on glycolysis for rapid ATP production do?
remove excess NADH from the cytosol and put it into the mitochondria to further speed of glycolysis
what does the cell do under conditions of excess?
it uses the mitochondria to supply the cytosol with:
excess citrate for synthesis of fatty acids and sterols
reducing power (such as NADPH) for biosynthesis using excess mitochondrial reducing power
where does the energy released during oxidative phosphorylation go? as in, what does it become?
it comes off as heat
what happens to the energy of the electron as it passes down the ETC? what about electron affinity?
it loses energy as it’s passed along the chain. transfer of electrons is in the direction of proteins with increasing electron affinity: the final electron acceptor is oxygen, which has the highest affinity for electrons
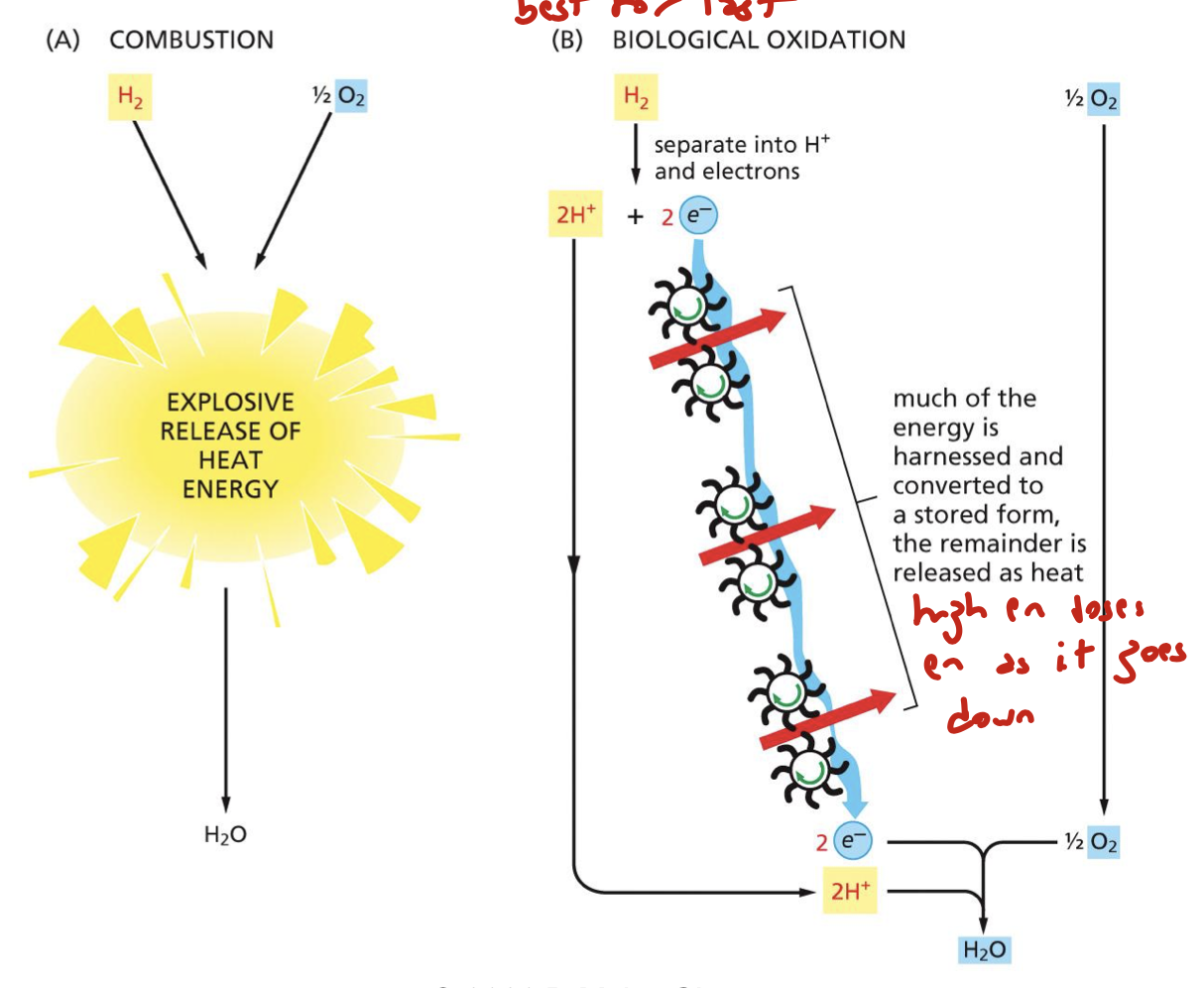
what are two reducing agents of the electron transfer chain?
NADH and FADH2
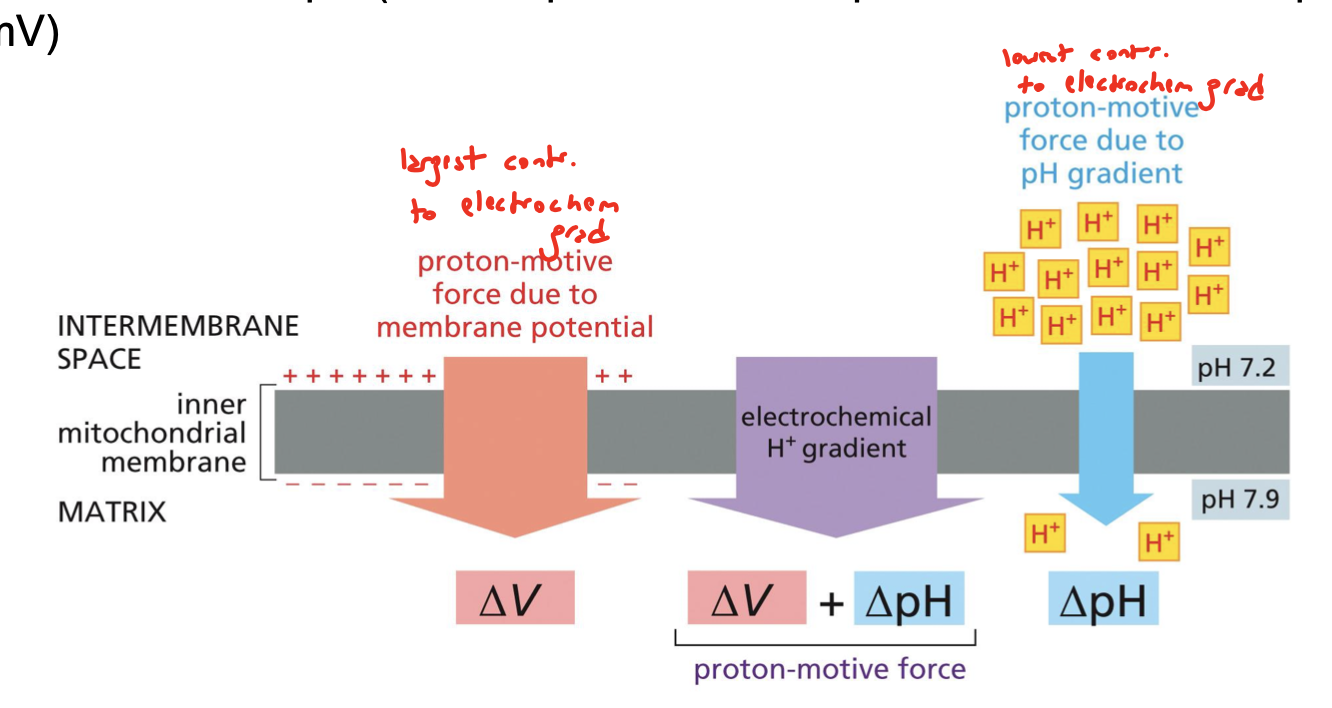
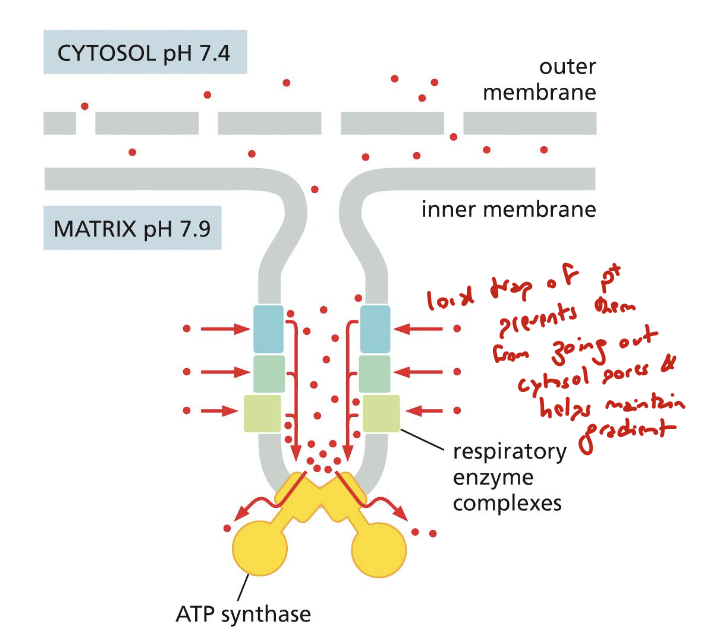
what is the pH of the IMS?
7.4
what is the pH of the matrix?
8/0
what two things represent proton motive force (PMF)?
pH gradient (matrix is more basic than the IMS) and the membrane potential of the electrochemical gradient of the mitochondria. this PMF tends to drive hydrogen back into the matrix.

what contributes most and least to the electrochemical gradient: membrane potential and pH gradient?
membrane potential (deltaV) contributes the most, pH gradient (deltapH) contributes the least
define redox potential with words and a mathematical definition
a measure of electron affinity: voltage difference between a standard redox pair and a second redox pair = redox potential
ex: electron moves spontaneously from low redox potential of NADH/NAD+ pair to high redox potential pair (O2/H2O)
another way to think of it: if you’re less electron-loving, you have low redox potential. high electron-affinity? high redox potential.
redox potential is designated as E’0 and is measured in mV
what is the electron donor:acceptor ratio of NADH:NAD+?
strong electron donor:weak electron acceptor
what (and how many) electron carriers would you see in the respiratory chain?
the RC will contain one or more of these carriers, which usually transfer one electron at a time:
cytochromes: heme (porphyrin) + apocytochrome (which you can detect via absorbance)
FeS clusters
flavin and copper atoms
these are also called prosthetic groups
what are examples of mobile electron carrier molecules?
cytochrome c: aqueous and transfers one electron at a time
ubiquinone (coenzyme Q): lipid soluble, can transfer 2 electrons at a time
what kind of element makes a good electron carrier molecule in the respiratory chain?
transition metals due to the fact that they have several different oxidation states with closely spaced redox potentials allowing them to accept or donate electrons readily
what (general term) techniques allow us to assay electron carriers in the respiratory chain?
spectroscopic and sepctrophotometric techniques
ex: absorbance is used to detect apocytochrome
rank these four redox electron carrier pairs in order from highest to lowest redox potential: UQH2/UQ (ubiquinone), H2O/O2, red cyt c/ox cyt c, NADH/NAD+
H2O/O2 > red cyt c/ox cyt c > UQH2/UQ > NADH/NAD+
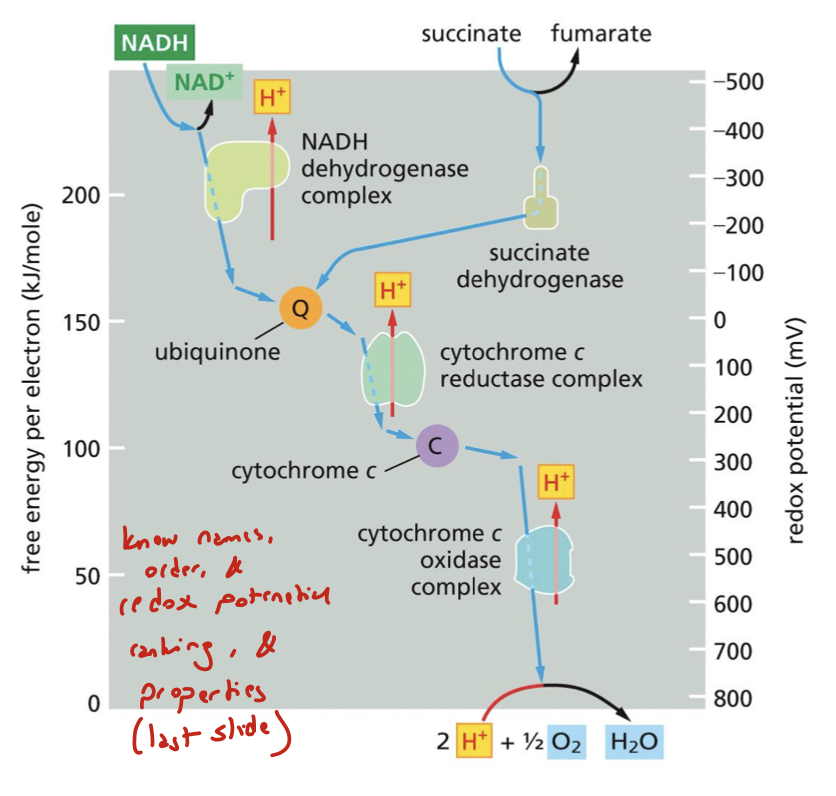
how can we view the redox pairs of the ETC?
in vitro by isolating them in liposomes in the presence of appropriate artificial electron acceptors and donors
what drives the energetics of the redox pair complexes in the ETC?
gaining and losing hydrogen protons and electrons
which complex is the first acceptor of FADH2 in the mitochondrial ETC?
it feeds electrons from Complex II to Q through C
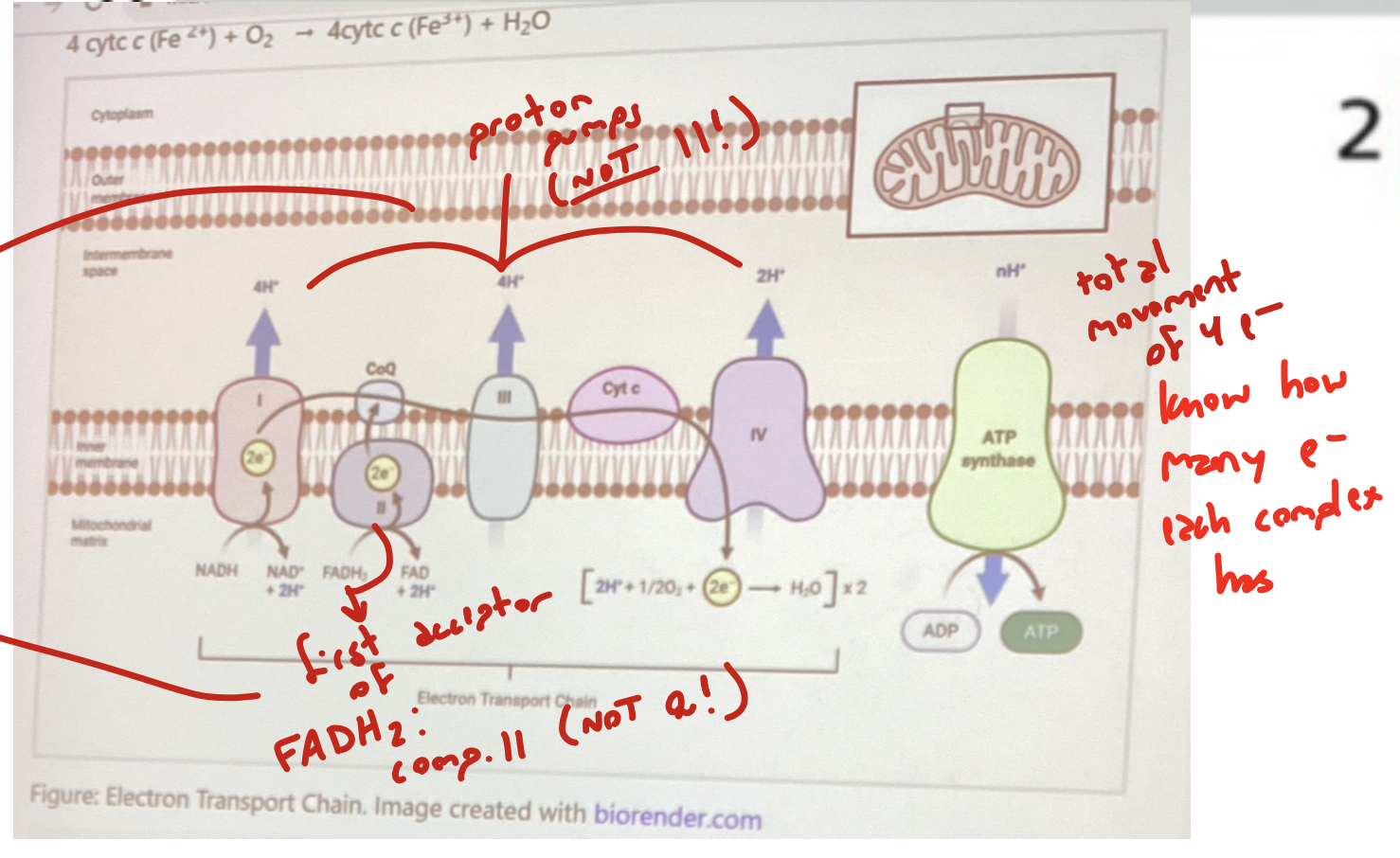
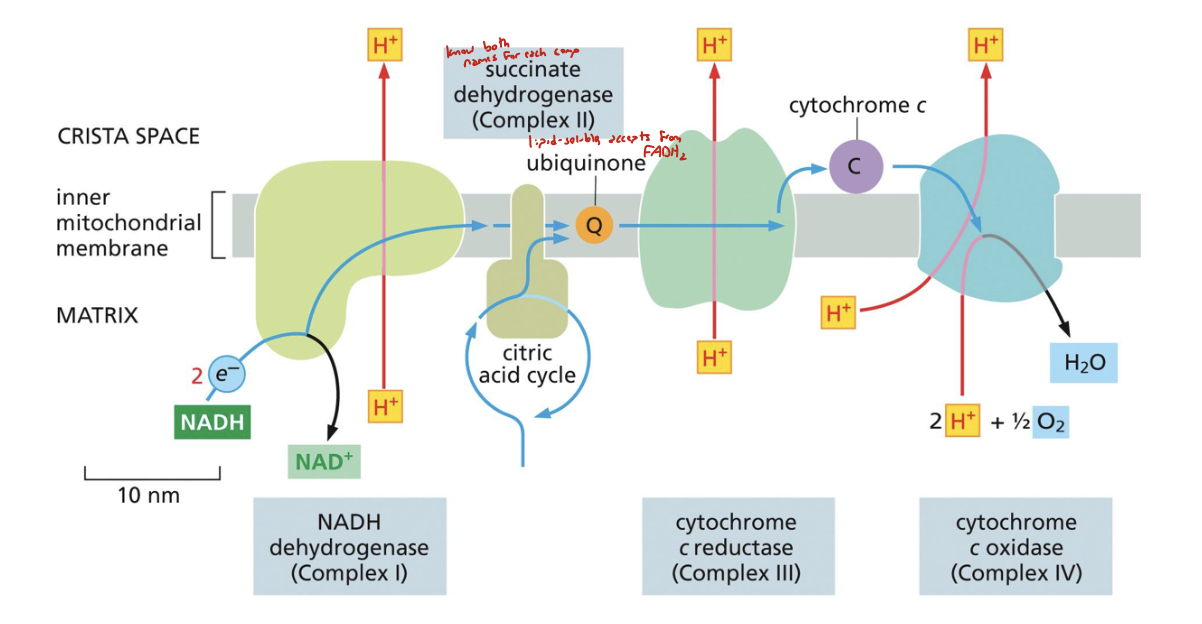
list the ETC complexes in order. when applicable, mention which are proton pumps; which prosthetic groups they have; how many electrons they contain
complex I: aka NADH dehydrogenase, a proton pump that contains flavin and FeS clusters as prosthetic groups. NADH:NAD+ passes two electrons through it into CoQ
complex II: aka succinate dehydrogenase (SDH), has FeS and hemes as prosthetic groups. doesn’t pump hydrogen. FADH2:FAD works here (citric acid cycle here!)
complex III: aka cytochrome c reductase. proton pump with hemes and FeS clusters as prosthetic groups. accepts electrons from CoQ and passes it to cyt c (specifically 2 electrons)
complex IV: aka cytochrome c oxidase (COX). terminal electron acceptor. proton pump with heme and copper as prosthetic groups. takes electrons from cytochrome c and passes it to H2O:O2 in the matrix
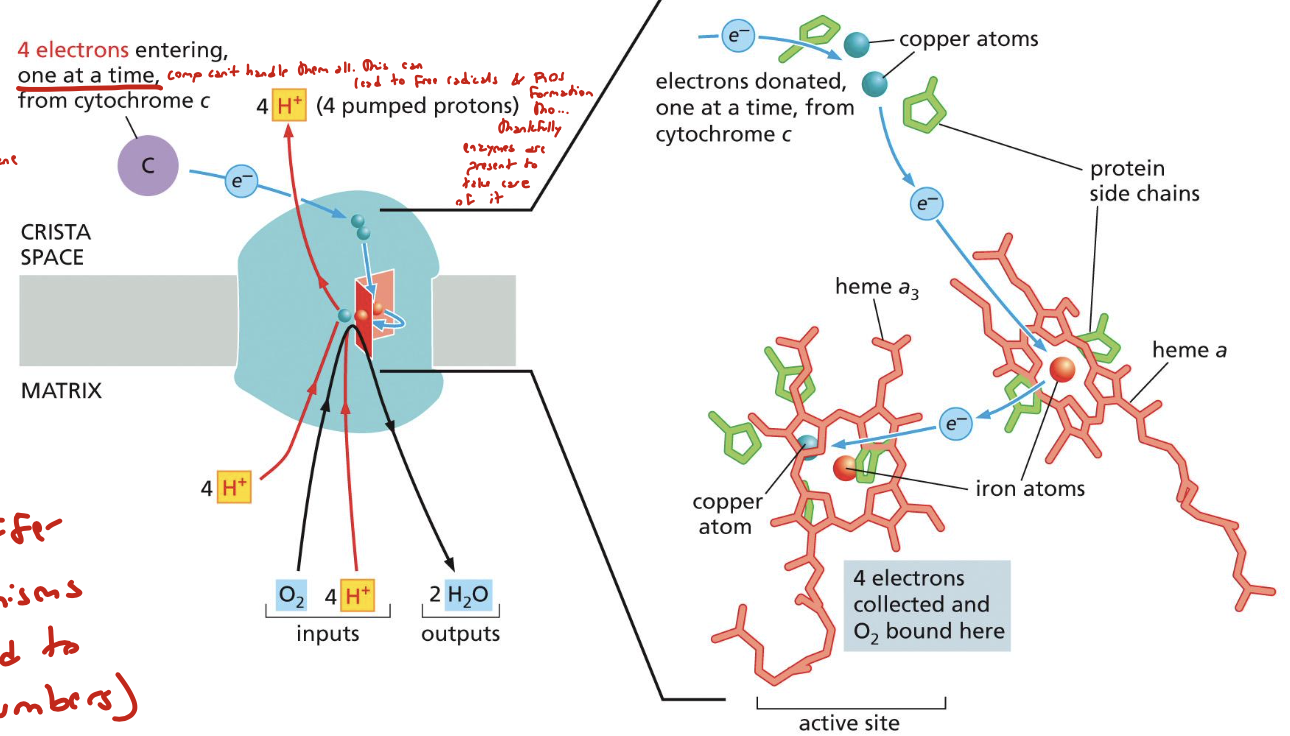
how many total electrons result from the ETC?
4
what does a high electron affinity in oxygen mean?
it means it releases a large amount of energy when reduced to water, but it can also produce ROS. solution? COX “clamps” oxygen until all 4 electrons are accepted so that it can be fully reduced and ROS is prevented
what holds COX to the mitochondrial membrane?
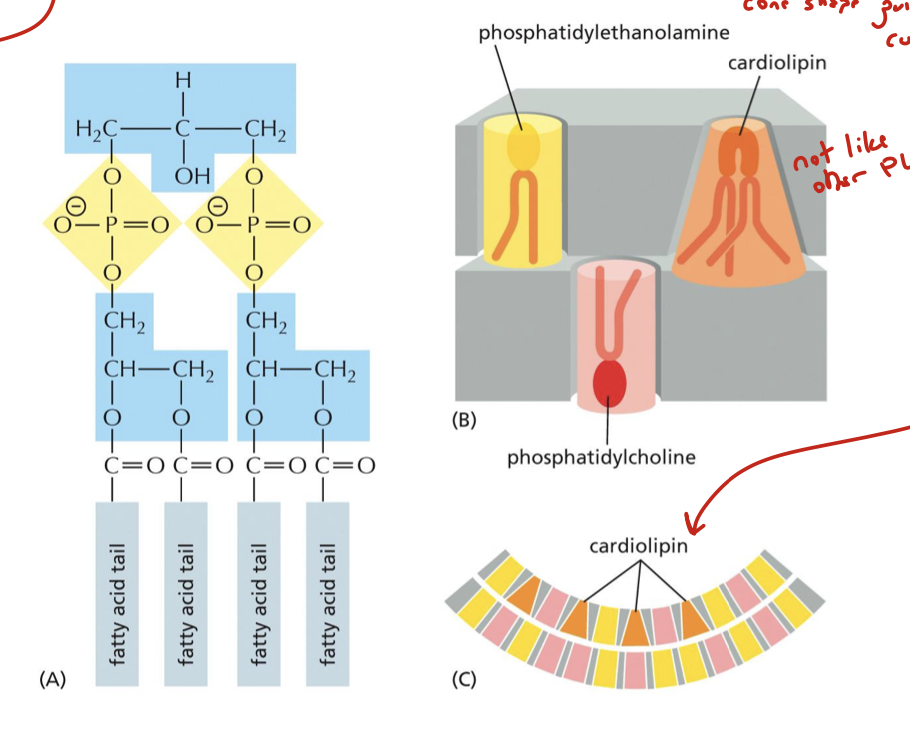
cardiolipin
are COX subunits conserved across organisms?
no. every organism has a different amount of subunits
how can we experimentally screen for mitochondrial mutants in yeast? why can we do this?
since yeast is a facultative anaerobe, it can generate ATP from glucose in the absence of oxygen or in the presence of a mitochondrial defect. so, strains that have these defects grow almost normally on glucose medium but fail to grow on ethanol or glycerol because these media contain non-fermentable carbon sources.
what complexes make up the respiratory chain supercomplexes? what should we know about their structure? how can we view/identify it?
all the proton pumps: complexes I, III, and IV
structurally, they overlap (hence “supercomplex”). cardiolipin holds them together (hydrophobic, basically lipid glue)
this supercomplex can be identified by cryoEM of gently isolated proteins found within it
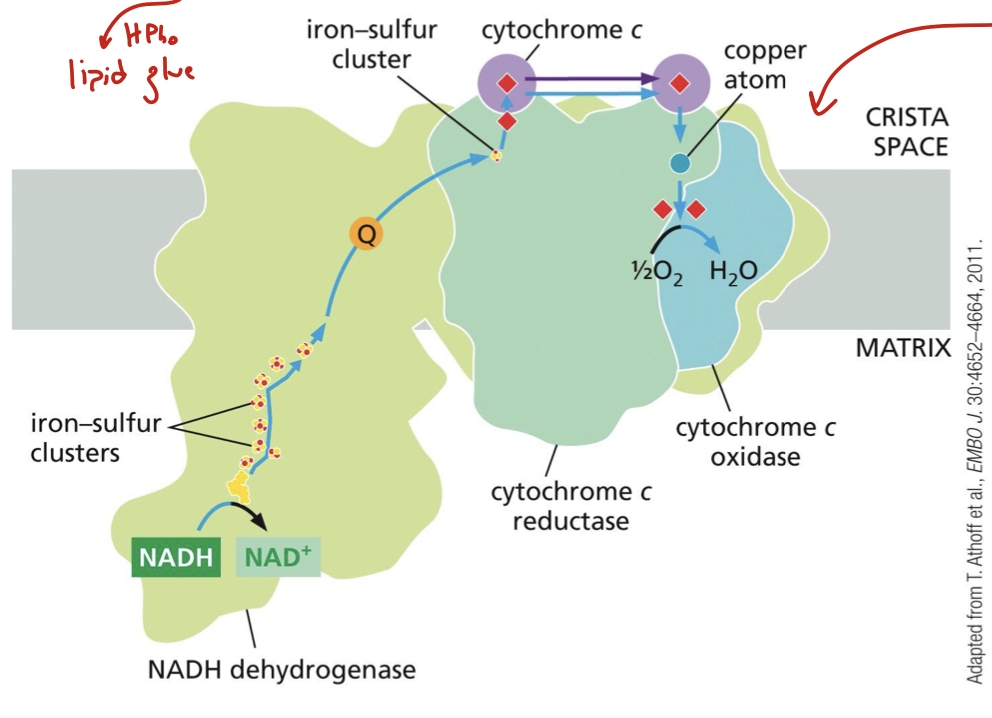
where is cardiolipin produced?
in the mitochondrial membranes, where it interacts closely with membrane proteins involved in oxidative phosphorylation and ATP transport
what does cardiolipin do in the cristae?
supports membrane curvature: hydrophobic lipid glue! it has a conical shape which adds to the curvature
what drives oxidative phosphorylation?
favorable ATP/ADP ratio: pool of ATP drives cellular processes like a battery, so we need more ATP than ADP
large negative free energy change from hydrolysis of the phosphate bond when [ATP] is high, so we need it high
what is F1F0 ATPase?
a proton-driven turbine that generates ATP. F0 is the rotor, F1 is the stator
it converts mechanical energy to chemical bond energy: allosteric shape changes couple ATP synthesis, an unfavorable reaction, to proton flow (energetically favorable)
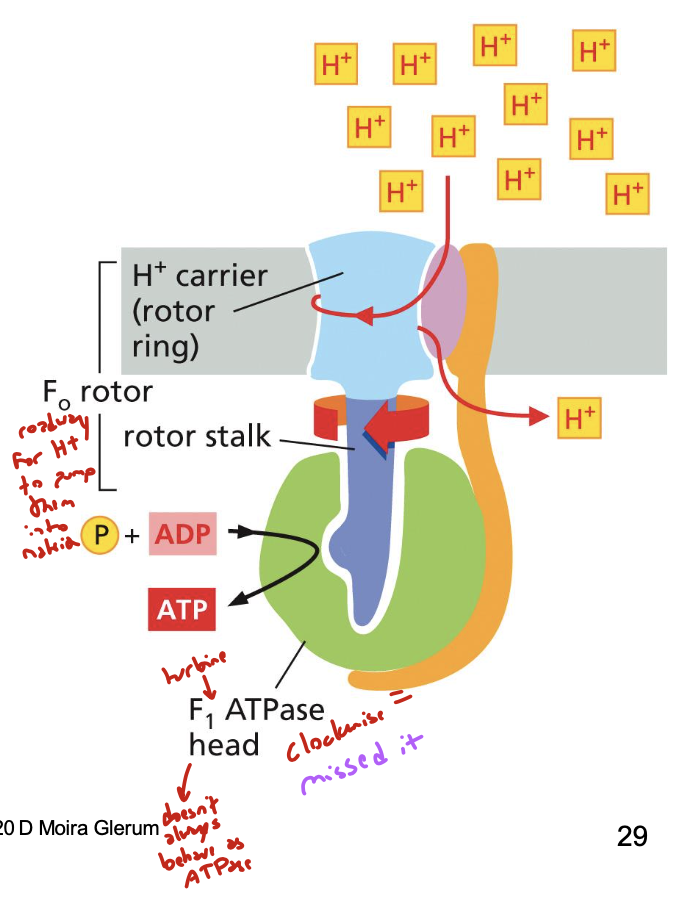
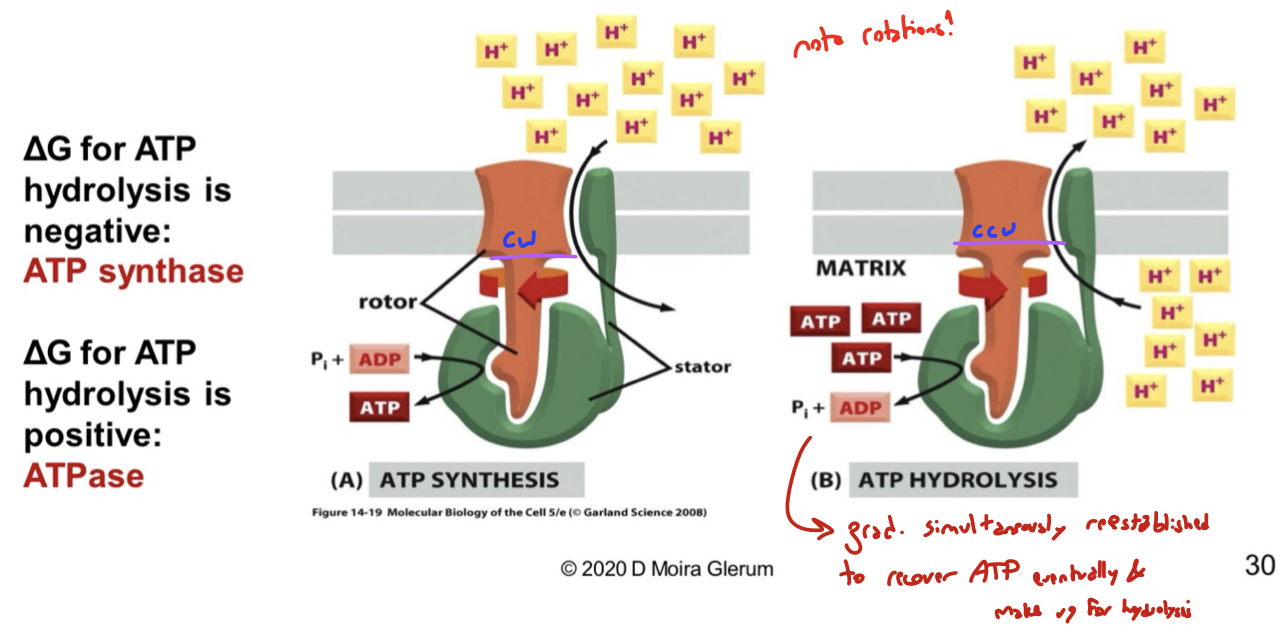
list the two ways F1F0 ATPase rotates and what each direction does
clockwise: performs ATP synthesis. protons are pumped into mito matrix. motor acts as an ATP synthase because deltaG for hydrolysis is negative
counter-clockwise: performs ATP hydrolysis. protons are pumped out of mito matrix. motor acts as an ATPase when deltaG for hydrolysis is positive
these directions are in equilibrium: the proton gradient is simultaneously re-established to recover ATP eventually and make up for hydrolysis
how do cristae help maintain electrochemical gradient?
by “trapping” protons to prevent them from leaving out of cytosolic pores
how do transports manage to bring metabolites across the IMM?
by coupling it with the favorable flow of protons
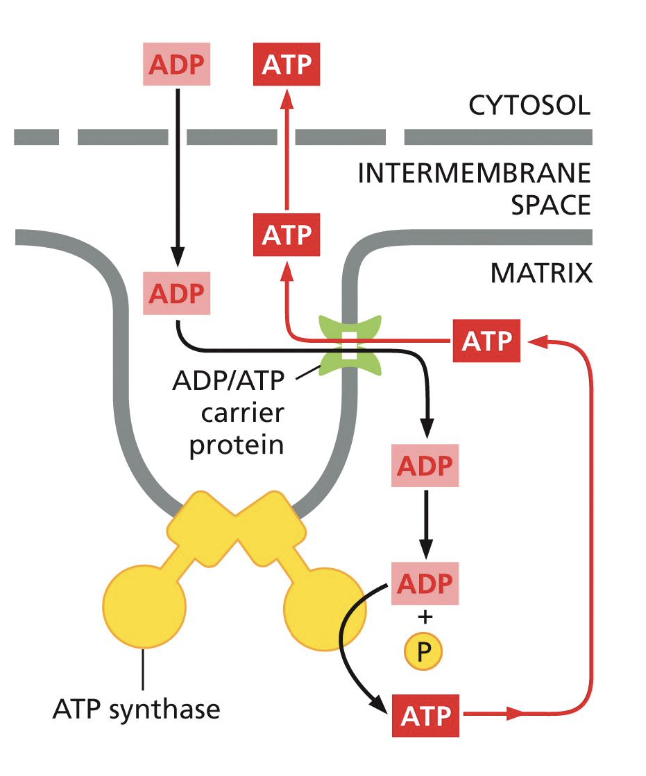
list and describe two ways the respiratory cycle is controlled
uncoupling agents (ex: DNP)
disconnect electron transfer from ATP production
lipid-soluble, dissipate PMF
causes an increase in oxygen uptake
brown fat
most energy of oxidation is dissipated as heat due to the presence of uncoupling protein. this is switched on when heat is needed
what kind of electron carriers do prokaryotes use to generate energy?
carbon, nitrogen, and sulfur instead of oxygen (anaerobes)
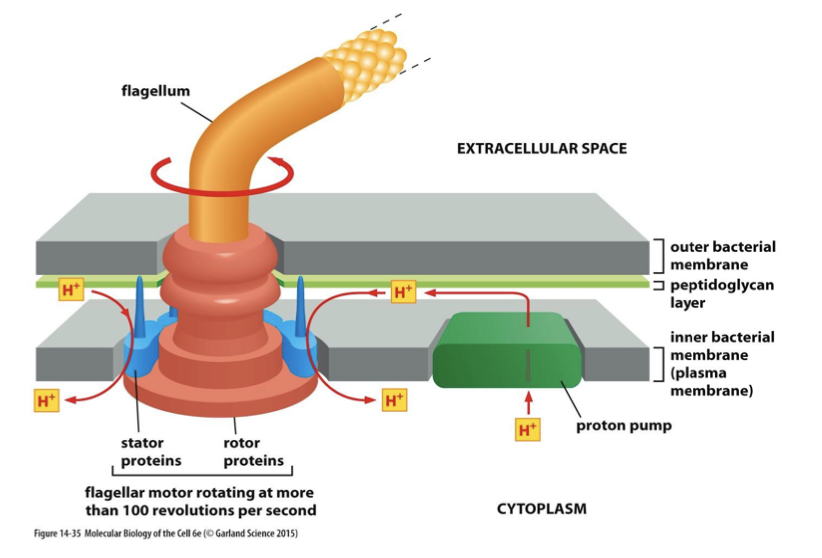
how does the rotor ATPase work in prokaryotes?
it maintains a proton gradient across the plasma membrane. in forward direction: aerobes. in reverse: anaerobes.
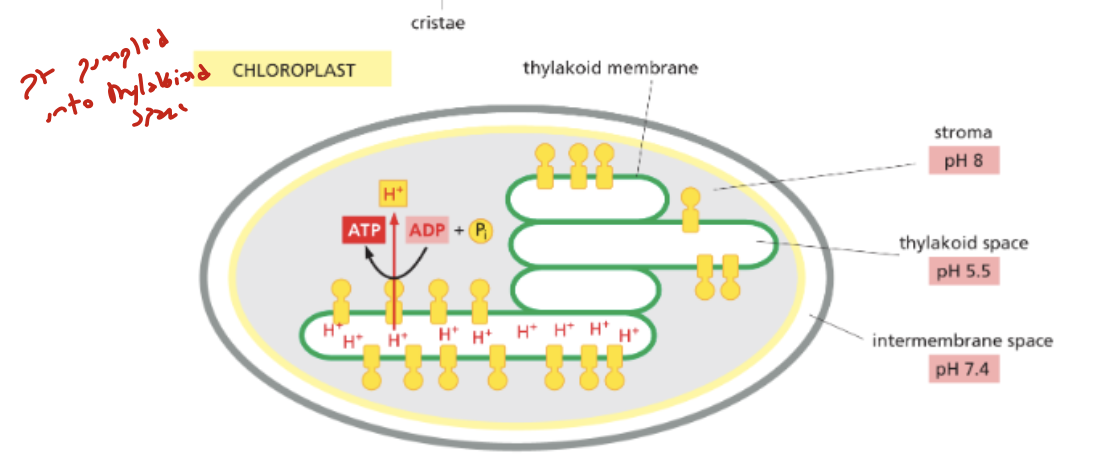
where are protons pumped in chloroplasts?
thylakoid membrane
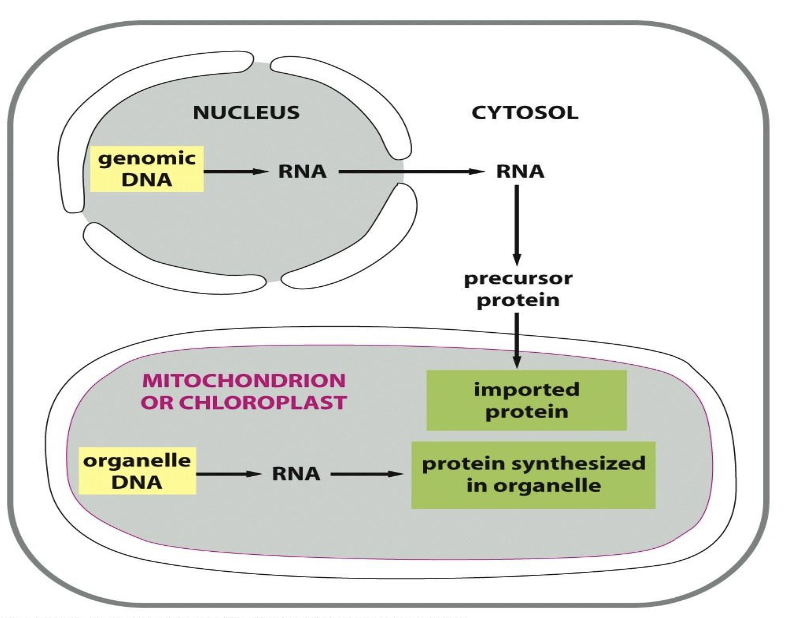
differentiate between nuclear and mitochondrial DNA
nuclear DNA:
is transcribed, translated, and imported via TOMs and TIMs
protein traffic is usually unidirectional
mitochondrial DNA (mtDNA):
expressed in the mitochondrion itself
all protein machinery for transcription and translation is nuclear-encoded though
note that plants have an additional genome for chloroplasts
what induces and coordinates fission of the mitochondria?
dynamin, aka Opa1
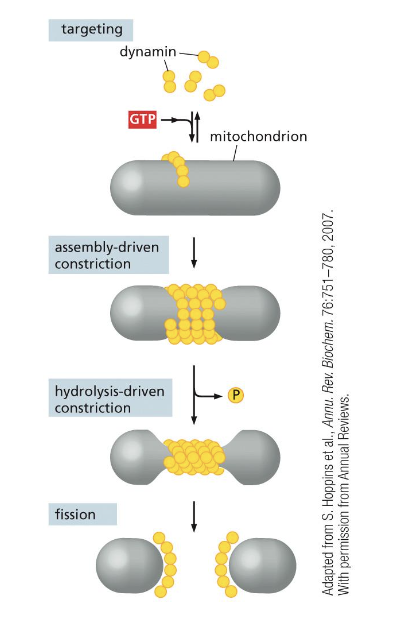
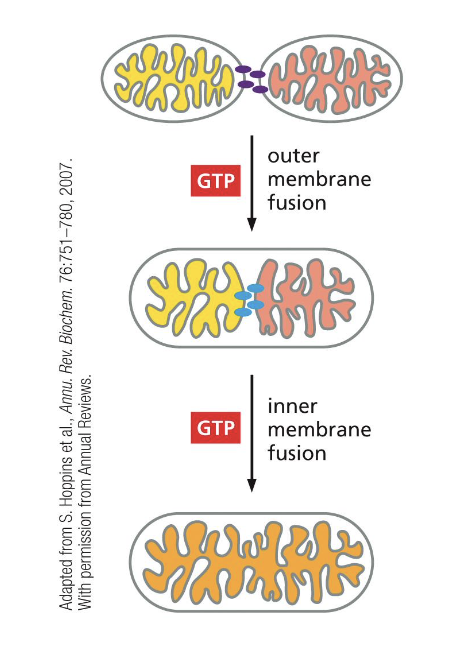
what coordinates the fusion of mitochondrial membranes?
GTPases in the IMM and OMM
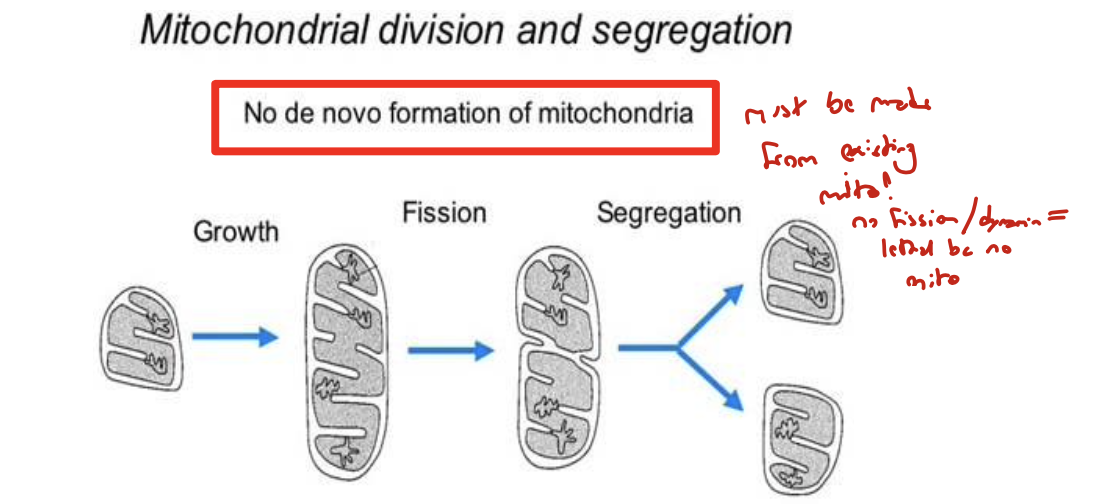
does the mitochondria have a life cycle it follows for fission?
not really. fission seems to happen independent of life cycle and may be tied to metabolic need instead
how many protein-coding genes does the mtDNA have?
13

are all mitochondrial complexes mtDNA-encoded?
no. only I, III, IV, and V are. II is nuclear DNA-encoded
which DNA encodes for the proteins for transcription and translation?
nuclear DNA. any transc, transl, repair, and packaging proteins are nuclear-DNA encoded and are not the same proteins as those involved in maintenance of the nuclear genome
does mtDNA have histones? why or why not?
no. but it has some other proteins associated with it
no histones bc it has prokaryotic origin (and prok have no histones!)
what genetic pattern is mtDNA inherited by in:
yeast
plants and animals
mammals
yeast: biparental fashion
plants and animals: uniparental
mammals: maternally inherited
why isn’t mammalian mtDNA inherited paternally?
sperm mtDNA is altered during transport toward the egg so it’s removed by autophagy to avoid the error-prone DNA (excess mito activity of the sperm to move the flagella, so more DNA replications are made which ups the mutation rates)
list 4 features of mtDNA
dense gene packaging: no introns in humans, but yeast has introns…
relaxed codon usage: uses less tRNA
variant genetic code: UGA is not the stop in animal mito (it is in plants though)
increased mutation rate (10x) compared to nDNA
DNA repair pathways are crude
exposure to ROS
replication is not controlled by a cell cycle: not just in S phase, not as controlled
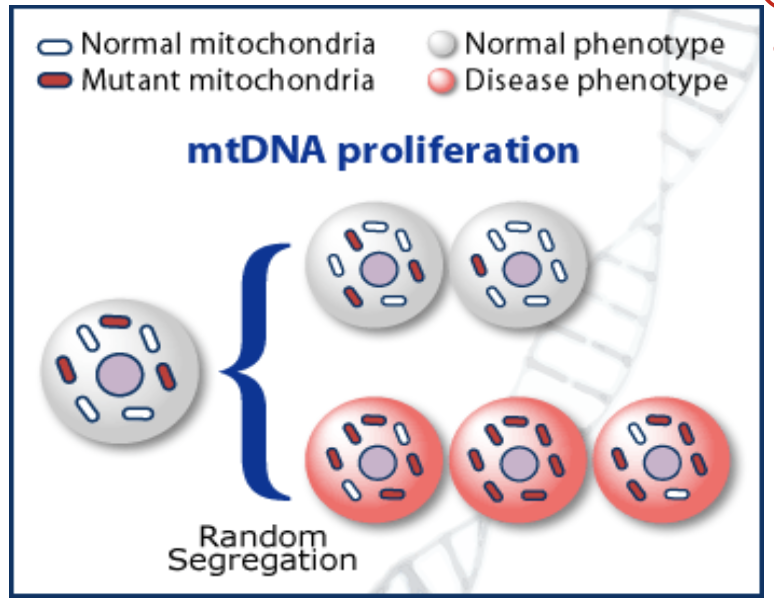
define heteroplasmy
not all mtDNA molecules in a mitochondrion or cell are the same and so we have mixed mtDNA populations. this is because there can be multiple mtDNA molecules per mitochondrion
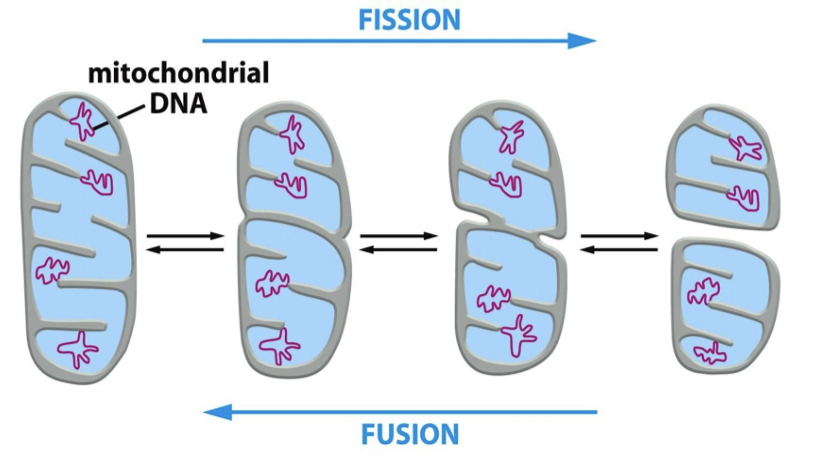
what does random segregation of mtDNA during cytokinesis lead to?
heteroplasmy and threshold effect
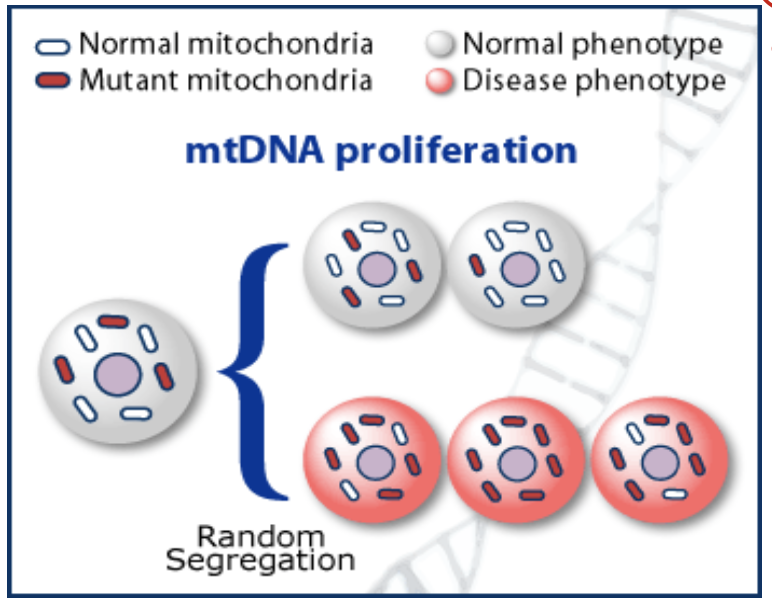
define threshold effect
a given mutant load does not exact the same phenotypic conseuqneces on different tissues.
ex: muscle cell vs skin cell
skin cell doesn’t use as much mito, so it has a higher threshold of defective mtDNA
muscle relies heavily on mitochondria, so it has a lower threshold for error