Genetic Principles in Blood Banks
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
Blood Group Systems
groups of antigens on the RBC membrane that share related serologic properties and genetic patterns of inheritance
Ex: ABO and Rh systems
Blood Group Genetics
Genetic material in DNA
contained in chromosomes in the nucleus of every cell
Genetic material is replicated by mitosis (somatic cells) or meiosis (gametes).
Immature RBCs (reticulocytes) loses its nucleus, as the cell matures.
Contains coding info for blood groups
Phenotype vs. Genotype
refers to the observable physical and functional traits of an organism vs the genetic makeup that determines those traits.
Phenotype
Physical (observed) expression of traits.
Determined by hemagglutination of RBC antigens using antisera.
Ex: No agglutination with anti-A or anti-B antisera indicates type O blood.
Genotype
Actual genetic makeup.
Determined by molecular techniques or family studies.
Ex: A person with phenotype A could have genotype A/A or A/O; family studies are needed to confirm.
Punnett Square
Used to predict the probability of an offspring’s genotype
Summarizes every possible combo of maternal and paternal alleles of a particular gene
Gene
Basic units of inheritance on a chromosome.
A locus is the site at which a gene is located on a chromosome.
Alleles are alternative forms of a gene, found at each locus.
Antigens produced by opposite alleles are antithetical (e.g., Kpa and Kpb antigens).
Multiple alleles at a single locus are considered polymorphic.
Inheritance: Recessive, Codominance, Dominance
Recessive: Gene is expressed only when inherited by both parents.
Codominant: Equal expression of two different alleles. Blood group antigens are codominant.
Dominant: Gene that is expressed over another gene.
Genes that do not express a detectable product are considered amorphic (e.g., O gene).
Mendelian Principles
applied to blood group antigen inheritance
Independent segregation occurs when one gene from each parent is passed to the offspring
Independent assortment is demonstrated when group antigenes from diff. chromosomes are expressed separately → MIXTURE of GENETIC MATERIAL
Mendelian Principles Exceptions:
Linkage and Crossing over
Linkage
Occurs when 2 genes that are close to each other are inherited together.
Each set of linked genes = HAPLOTYPE
linkage disequilibrium.
Linkage disequilibrium
Haplotypes tend to occur at a higher frequency than for unlinked genes
Crossing Over
when 2 genes on the same chromosome combine and produce 2 new chromosomes.
Chromosomal Assignment: Xg Significance
Most blood group system genes are on autosomes, except for those of the Xg system.
Xg genes are found on the X chromosome.
If the father carries the Xg allele, he will pass it to all of his daughters but not to any of his sons.
If the mother carries the Xg allele (not the father), all of their children will express Xg.
Xg System
Xg genes are on the X chromosome. Fathers pass the Xg allele to daughters only; mothers pass it to all children.
Heterozygosity and Homozygosity
A person who inherits identical alleles is called homozygous.
Examples: AA, BB, MM (M+ N–)
A person who inherits different alleles is called heterozygous.
Examples: AO, AB, MN (M+ N+)
Dosage
variation in antigen expression due to the number of alleles present.
In some blood group systems, persons homozygous for an allele have a “double dose” of an antigen on their RBCs compared with those who are heterozygous for an alleles
Homozygous expression of some antigens will show stronger agglutination compared with antigens that are heterozygous.
Genetic Interaction
location of inherited genes affect the expression of the antigen.
Alleles on the same chromosome are cis to one another.
Alleles on opposite chromosomes are in the trans position.
Phenotype Calculations Ex 1: A patient with multiple antibodies (anti-C, anti-E, anti-S) needs blood.
70% are C positive, 30% negative.
30% are E positive, 70% negative.
55% are S positive, 45% negative.
Calculation: 0.30 x 0.70 x 0.45 = 0.0945 or 10%.
About 10% of the population will be negative for all three antigens; about 1 in 10 units will be compatible.
Phenotype Calculations Ex 2: A patient with multiple antibodies needs 2 units.
66% are Fya positive, 34% negative.
72% are Jkb positive, 28% negative.
9% are K positive, 91% negative.
Calculation: 00.34 × 0.28 × 0.91 = 0.087 or 9% negative (9 out of 10)
2 units needed / 0.09 (antigen negative frequency) = 22.
Conclusion: Antigen typing of 22 units may be required to find 2 compatible units.
Hardy-Weinberg Formula
Basic Formula:
p + q = 1
(p + q)^2 = 1.0 or p^2 + 2pq + q^2 = 1.0
Where:
p = frequency of allele A
q = frequency of allele a
Hardy-Weinberg Formula Example: What is the frequency of q if p is 0.3? What are the genotype proportions?
q frequency: 1 – 0.3 = 0.7
AA = p^2 = 0.09 (homozygous for A)
Aa = 2pq = 0.42 (heterozygous for Aa)
aa = q^2 - 0.49 (homozygous for a)
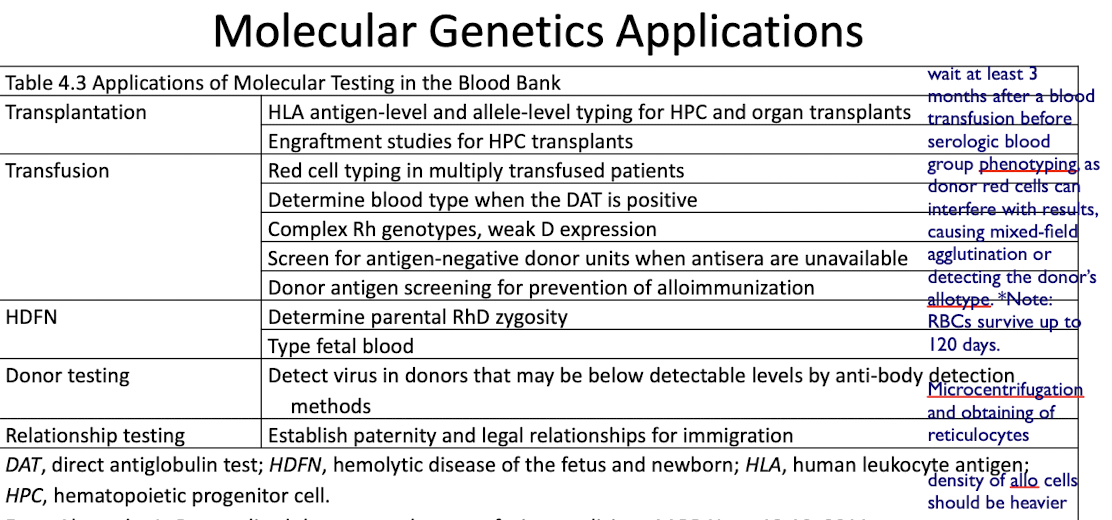
Transplantation
HLA antigen-level and allele-level typing for HPC and organ transplants
Engraftment studies for HPC transplants
Transfusion
Red cell typing in multiply transfused patients
Determine blood type when the DAT is positive
Complex Rh genotypes, weak D expression
Screen for antigen-negative donor units when antisera are unavailable
Donor antigen screening for prevention of alloimmunization
HDFN
Determine parental RhD zygosity
Type fetal blood
Donor Testing
Detect virus in donors that may be below detectable levels by anti-body detection methods
Relationship Testing
Used to Establish paternity and legal relationships for immigration, where unique red blood cell antigens or other cellular markers are inherited and analyzed to confirm biological relationships.
Polymerase Chain Reaction (PCR)
PCR rapidly and precisely multiplies specific DNA sequences.
DNA is denatured.
A primer is added to attach specific areas of DNA.
DNA is amplified and replicated.
PCR-Based HLA Typing Procedures
Sequence-Specific Primers (SSPs)
Sequence-Specific Oligonucleotides (SSOs)
Sequence-Based Typing (SBT)
Short Tandem Repeats (STRs)
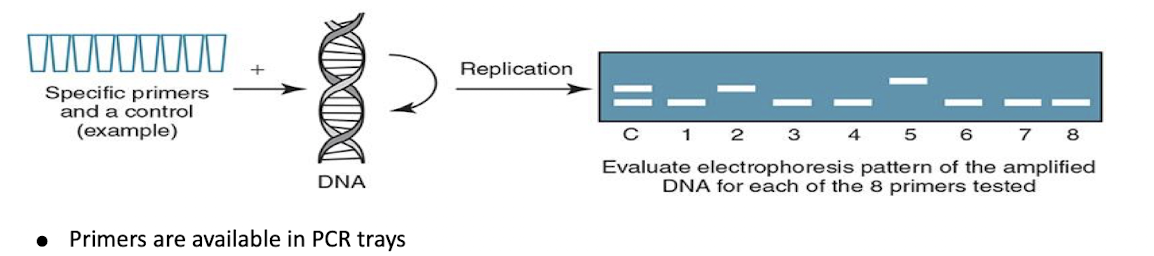
Sequence-Specific Primers (SSPs)
Primers are available in PCR trays.
Low resolution—identifies antigen level.
High resolution—defines specific alleles for the antigen.
Amplified DNA (amplicons) are assessed using gel electrophoresis.
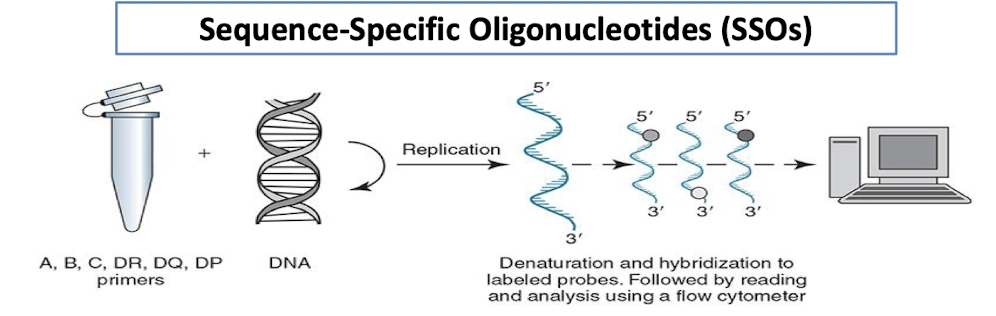
Sequence-Specific Oligonucleotides (SSOs)
A primer for each locus is used.
A, B, C, DR, DQ, DP (in separate wells)
A DNA probe allows the hybridized solution to be read and analyzed by a flow cytometer.
Sequence-Based Typing (SBT)
Provides high-resolution, allele-level typing.
Primers are similar to SSOs.
An instrument analyzes the nucleotide and amino acid sequences that correspond to the allele.
Short Tandem Repeats (STRs)
short sequences of DNA amplified to determine the % of engraftment in chimerism evaluation.
Donors and recipients have closely matched HLA alleles but may have slight variations in the DNA sequence called polymorphisms.
Chimerism evaluation uses these differences to determine the percentage of DNA from the donor in a stem cell recipient.
Molecular Applications of RBC Typing
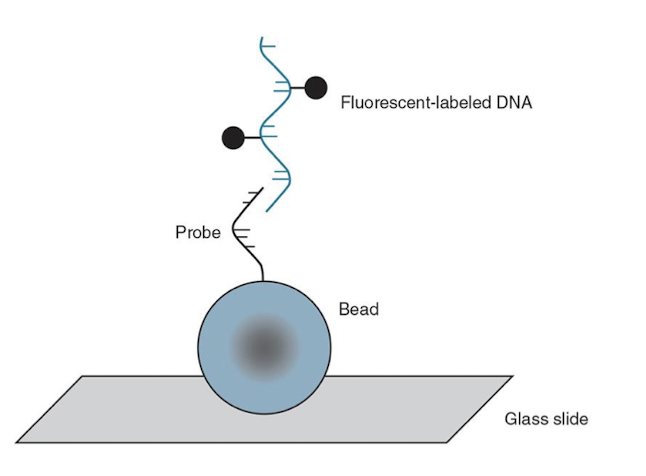
BeadChip Technology
Uses oligonucleotide primers attached to colored silica beads on a substrate (slide).
Amplified and digested DNA in question binds to the primers.
Computer analysis determines which primers have attached.