Finding the genes in a genome
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
what is the function of genes
region that is transcribed (includes 5’ & 3’ UTRs)
codes for proteins or RNAs
what is the function of cis-regulatory elements
enhancers, silencers promotors just upstream of the gene
regulates when transcription is initiated or not
what is the function of trans-regulatory elements
located far way from the gene of interest
non-coding RNAs that influence gene expression
what are repetitive regions
transposons
retrotransposon
single sequence repeats
what are transposons
a repetitive gene region capable of moving from one region to another
may require specific sequences of nucleotides to move
movement is called “transposing”
what is the functional importance of transposons
cause mutations (via insertion)
alter gene expression
induce chromosome rearrangment
what are retrotransposons
a repetitive gene region which must be reverse transcribed into DNA before inserting into a new region
moves similar to transposons
what is the functional importance of retrotransposons
causes mutations (via insertions)
alter gene expression
induce chromosome rearrangements
what are single sequence repeats
regions of genome composed of tandem repeats of bases
may have structural/functional properties
high rate of mutations allow number of repeats to contract/expand
what does each nucleotide contain
sugar phosphate backbone
a deoxyribose sugar
nitrogen containing base
what is the template strand
the DNA strand transcribed by RNA polymerase to make RNA
we read DNA 5’ - 3’
what is the coding strand
the DNA strand that has the same sequence as the RNA coded by the template strand (T rather than U)
what are some ways to analyze a genome sequence
identify genes from:
open reading frames
transcriptional & translational control regions (promoters, terminators, ribosome binding sites)
after identification:
determine aa sequence
identify intron/exon boundaries
identify gene variation (mutation)
how many reading frames are in DNA
six reading frames
3 for “top strand”
3 for “bottom strand”
how can genes be found in the genome sequence
can be found from:
open reading frame (start & STOP codons)
promotor regions
enhancer & silencer regions
translational control elements in 5’ UTR
how does a microarray work
an RNA sample is converted into cDNA
cDNA is exposed to transcript probes which are complementary to specific sequences
they bind and reveal the identity of the cDNA
what are some disadvantages to microarray
low sensitivity
low dynamic range
known transcript only
how does RNA sequencing work
an RNA sample is converted into cDNA
samples are aligned and a sequence is revealed
what are some advantages to RNA sequencing
high sensitivity
high dynamic range
novel transcripts sequences identified
what is the difference between a reading frame & open reading frame
reading frame - one of the 6 different ways to read codons
open reading frame - a DNA sequence that occurs between a start codon and STOP codon
must be >=30 nucleotides
what is the difference between an open reading frame & “real” open reading frame
open reading frame - may/may not code for a protein
“real” open reading frame - region between start & stop codon, that produces a real protein
sometimes RNA
what is defined as the “real” open reading frame in eukaryotics
the removal of all intron and splicing of the exon together to produce a sequence of codon that will code of a protein
what is a difference between searching for open reading frames in prokaryotes & eukaryotes
prokaryotes - no introns, hence we look for long sequences of codons unbroken by STOP codons
eukaryotes - introns are present hence we need to find the boundaries between them and exons, then splice them out and then search for the open reading frames.
what is a splice site
the region between an intron & exon, removed by splicosome
3’ splice sites: CAG|G
5’ splice sites: MAG|GTRAGT
NOTE: M = A or C & R = A or G
what are transcript variants
alternate use of intron/exon boundaries within genes, this means multiple mRNA transcripts can be produced from a single gene
each variant is a different open reading frame
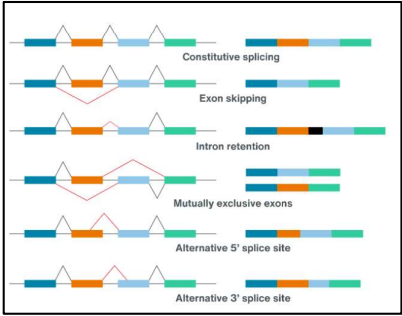
what is alternative splicing
refers to the different ways in which a spliceosome can remove introns (even exons) during mRNA modification
what is homology searching
if a paralogue (same species) or orthologue (different specie) has been identified for the gene you’re searching
existing info can help identify intron/exon boundaries
how can reverse transcriptase (PCR) find RNA
design primer based on predicted sequences of gene regions
if gene exists/expressed, PCR primers will bind to RNA transcript & create cDNA copy
detection of cDNA copy validates that predicted gene is “real”
how can transcriptomics find RNA
extract RNAs from a cell sample
run transcriptomics experiment to detect all different RNAs present
detection of cDNAs or RNAs that correspond to the sequence of the predicted gene validates them as “real”
how can proteomics find proteins
extract proteins from a cell sample, denature & unravel & treat with protease
run a mass spectrometry to confirm identity
detection of proteins through proteomics with the same sequence as predicted proteins validates that predicted gene as “real”
how can homology modelling be used to find proteins
based on predicted protein sequences from predicted genes, search against a database of all known & validated proteins
when a high match is found, this provides strong evidence that the predicted protein is a real protein
how can a western blot find proteins
design an antibody that recognises a part of the protein sequence your searching for
extract proteins from cell sample, then denature and add antibody (second antibody as indicator)
run a western blot to determine if protein has been bound to by antibody
detection to protein validates that protein isoform exists