Enzyme kinetics and inhibition
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
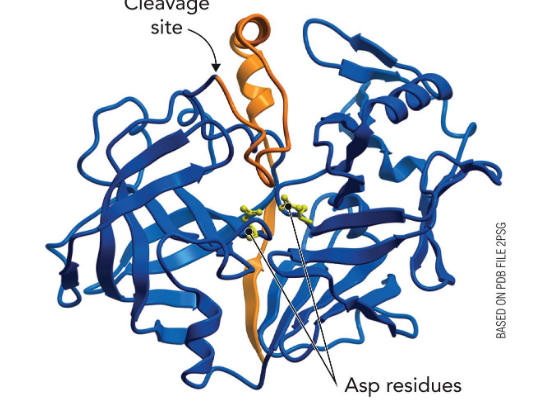
HIV Protease
Dimeric “Aspartyl Protease”, two identical subunits w/ active site ASP residues, flaps close and structure rigifies, major drug target of anti virals
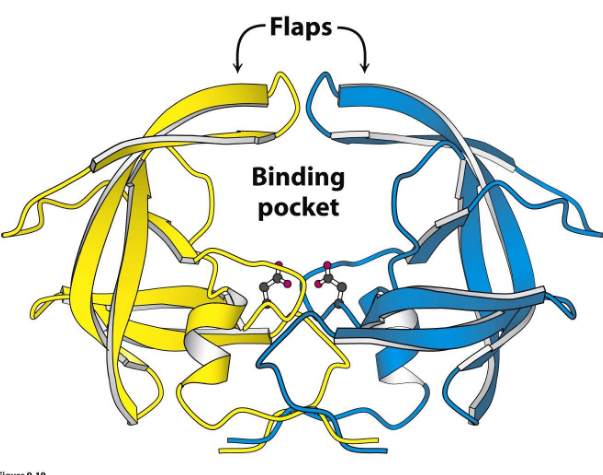
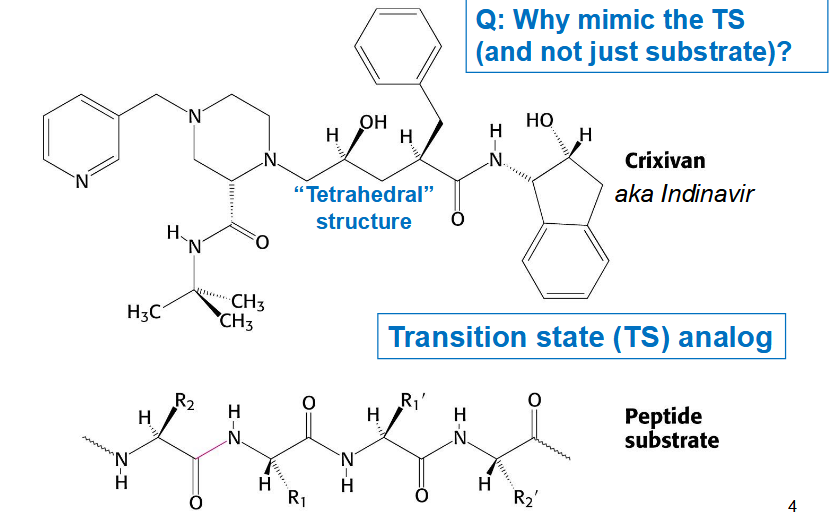
HIV protease inhibitors
Indinavir that mimics transition state: prefers transition state (maximum binding interaction
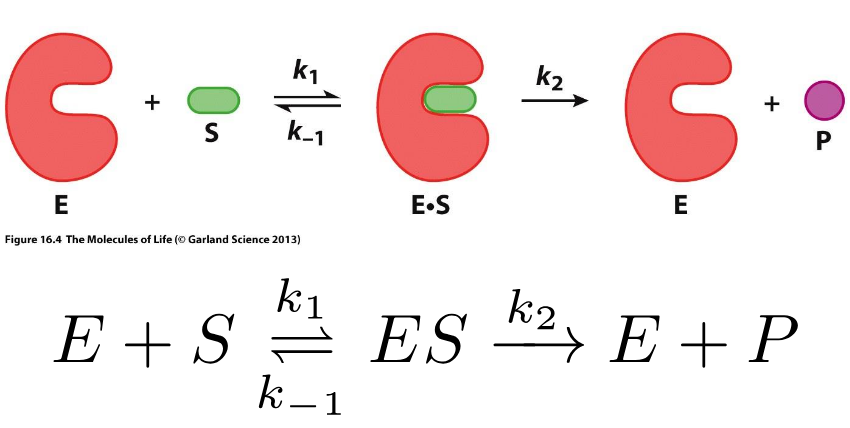
Enzyme Kinetics
Michaelis-Menten graph
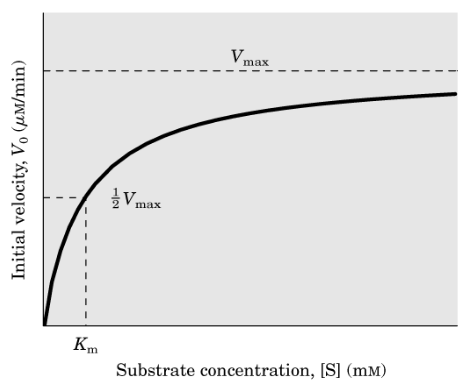
Michaelis Constant (Km)
Ratio of rate constants for elementary reaction steps, strength of ES complex formation
for rapid equilibrium Km = Kd
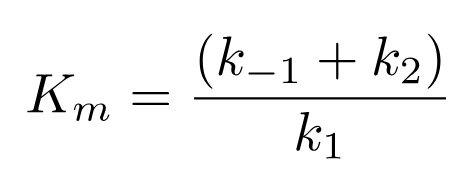
Michaelis-Menten Assumptions
[enzyme] does not change
k2 >> k-2 (no back reaction, easy at early time points)
[E]total << [S] (excess substrate!)
[S] does not change much with time
Steady-state of [ES]
Steady-State of the Michaelis complex [ES]
production and consumption of the ES complex proceeds at the same rate, the concentration of ES is constant
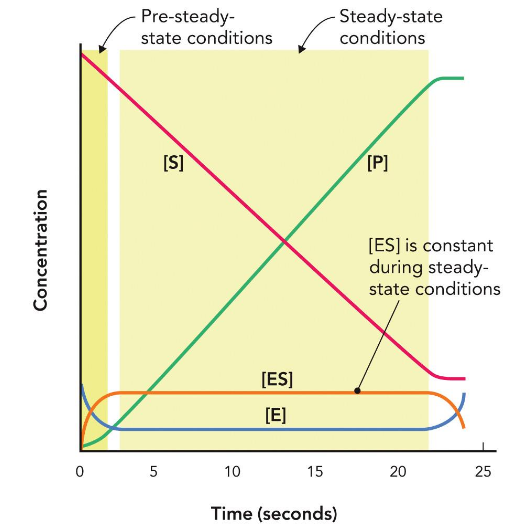
Monitoring Kinetics under Steady-State Conditions
enzyme is binding to substrate → converts to product → unbinds → binds to new substrate
Michaelis Menten Equation
double the enzyme concentration → Vmax doubles, but Km does not change
V0 is half-maximal at a concentration of S that is equal to Km
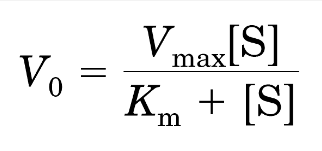
Saturation effect
observed in enzyme catalysis when plotting the initial velocity (Vo) against the substrate concentration([S])
Initial velocity (Vo)
measured at the beginning of the enzyme-catalyzed reaction, when substrate concentration can be considered constant, will decrease as the reaction progresses.
low concentrations of substrate
Vo increases almost linearly with an increase in [S]
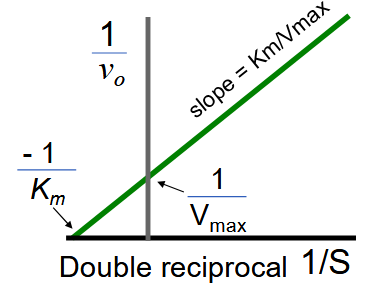
Lineweaver-Burk (Double Reciprocal) Plot
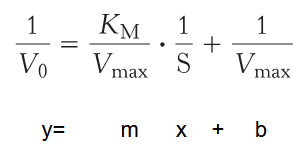
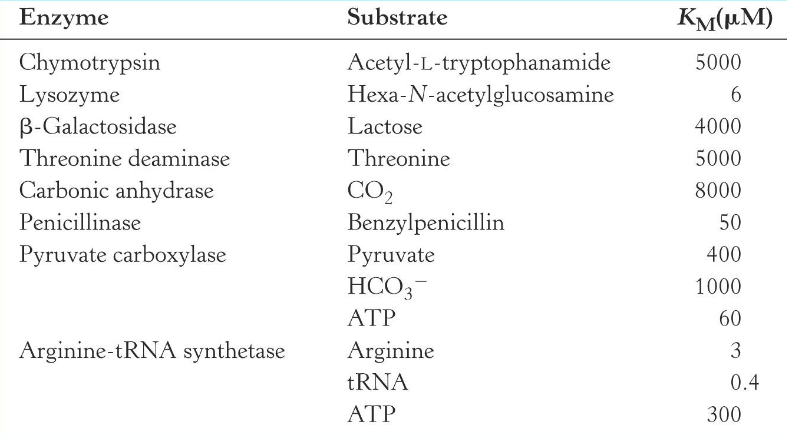
Km of some enzymes
lower the Km, the less Substrate the enzyme requires, which is associated with more efficient catalysis
most Km values range between 10-7 to 10-1 M
catalytic rate constant/turnover number (k2/kcat)
best, most efficient, enzyme has a maximal value of kcat
Catalytic Efficiency
V0 = kcat/Km [E]T [S]
Defines the efficiency of the enzyme Higher values, more efficient
![<p>V0 = kcat/Km [E]<sub>T</sub> [S]</p><p>Defines the efficiency of the enzyme Higher values, more efficient</p>](https://knowt-user-attachments.s3.amazonaws.com/6fdfe6bc-de68-4057-b027-313aee14ff12.png)
catalytic perfection
kcat/Km approaches diffusion limitted maximum (in water) ~ 108-109 M-1 sec-1

Bioavailablity
how much enzyme is around to ctalayse reactions; controlled by biochemical processes involved in protein synthesis and localization
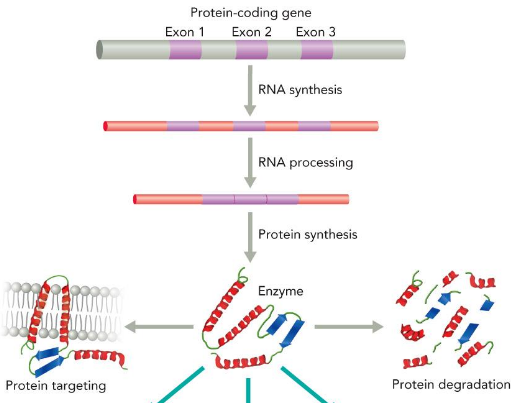
Catalytic Efficiency (regulation)
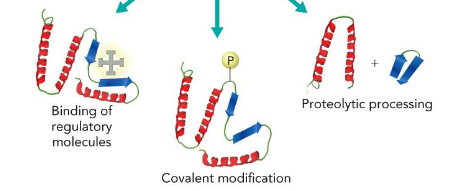
determined by protein modifications; Binding of regulatory molecules, Covalent modification, Proteolytic processing
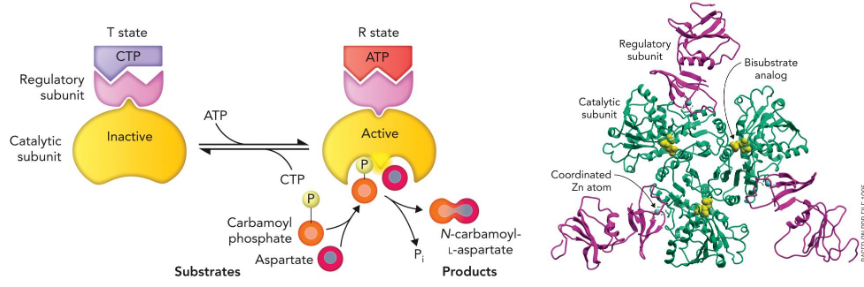
Allosteric inhibition
“feedback inhibition”
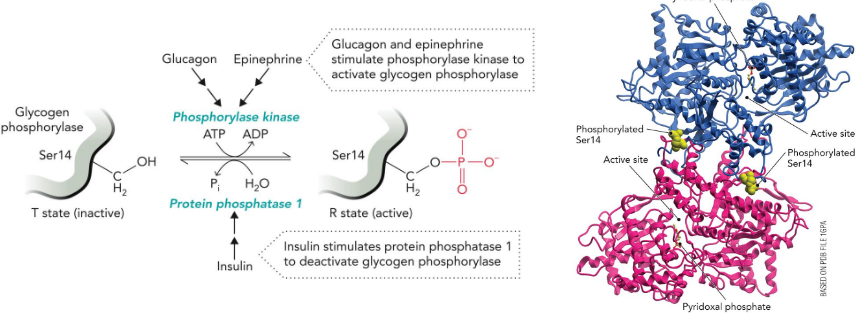
Phosphorylation
Proteolytic cleavage
Pepsinogen is a protease “zymogen” that is activated by autocleavage
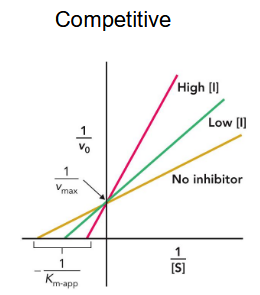
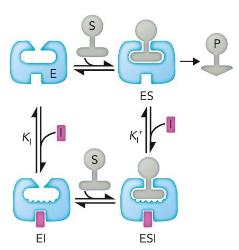
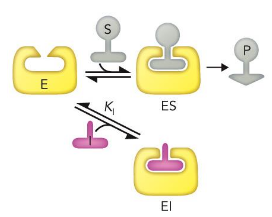
competitive inhibitor
binds at the active site (usually) and thus prevents the substrate from binding
affects Km but not Vmax; high levels of substrate relieve inhibition
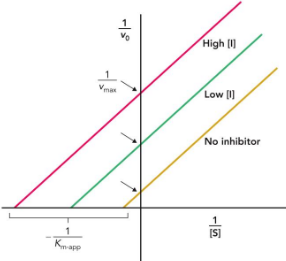
uncompetitive inhibitor
binds only to the enzyme–substrate complex
decreases Km and Vmax; cannot be overcome at high concentrations
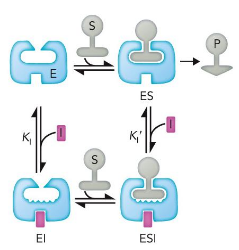
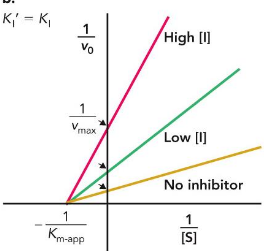
Noncompetitive/Mixed inhibitors
bind to both the enzyme and enzyme-substrate complex
Km unchanged; Vmax unattainable; reduces total enzyme concentration