4L6&7 Eukaryotic Transcription
1/62
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
63 Terms
eukaryotic gene organization
each gene has a promoter(s) and terminator(s)
exon = included, intron = excluded in primary transcript
overall expression of functional protein is regulated at
transcription
rna processing
mRNA turnover
translation
posttranslational modification
cellular trafficking
protein turnover
chromatin
eukaryotic chromosomal material of DNA, proteins (histones), and RNA
amorphous in G0 and interphase,
chromatin remodeling
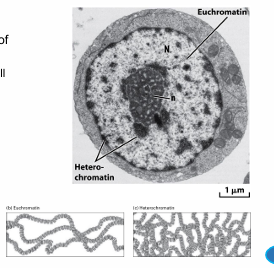
heterochromatin v euchromatin
hetero = more condensed, inactive, 10% chromatin, often in structures, dark staining
eu = normal, transcription, light staining
nucleosome
basic unit of chromatin contains 8 histones to make up histone core
in euchromatin, nucleosomes look like beads on a string
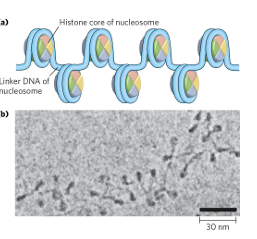
10nm fiber
nucleosome beads on a string (10nm is diameter of nucleosome)
wraps around histone twice
histones
positively charged,
DNA-histone core contacts are sequence independent
form electrostatic interactions and H bonds with negative backbone
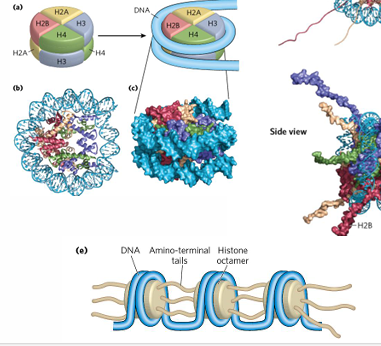
histone core
eight histone molecules, 2 copies of H2A, H2B, H3, H4
one core per nucleosome
histone H1
locks DNA to nucleosome
tails of histones
play key role in forming contacts between nucleosomes,
most of histone modifications here
epigenetics
chromatin structure is a product of epigenetic marks (histone mods and DNA methylation) and trans-acting transcription factors
cannot change DNA (not mutations)
histone tails
histone tail has H3 and H4 in N termini,
N and C termini of H2A H2B,
post translationally modified
histone tail modifications
acetylation = in areas of open chromatin, HAT acetylates lysine ***(Ac = activate, M = masks)
hypermethylation = often associated with closed chromatin, HMT methylates lys arg
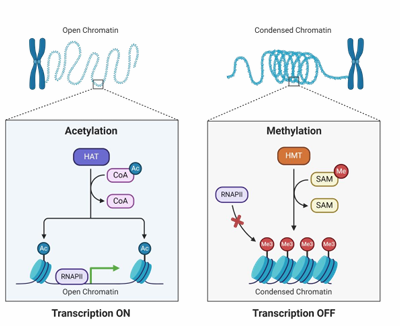
HAT
histone acetyltransferase, acetylates lysine in histone tails
HDAC
histone deacetylase removes acetyl groups from histone tails
HMT
histone methyltransferase methylates lysine and arginine in histone tails
histone acetylation
neutralizes positive charge on lysine in histone tails
reduces electrostatic nucleosome-DNA attraction
helps recruit chromatin remodeling complex
helps transcription activation
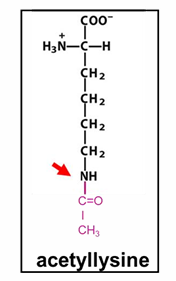
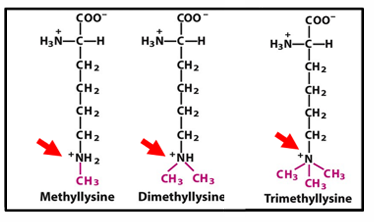
histone methylation
methylation retains positive charge on lysine/arginine
can activate or silence, typically closed chromatin
bind sites for enzyme
transcribed genes are
more diverse
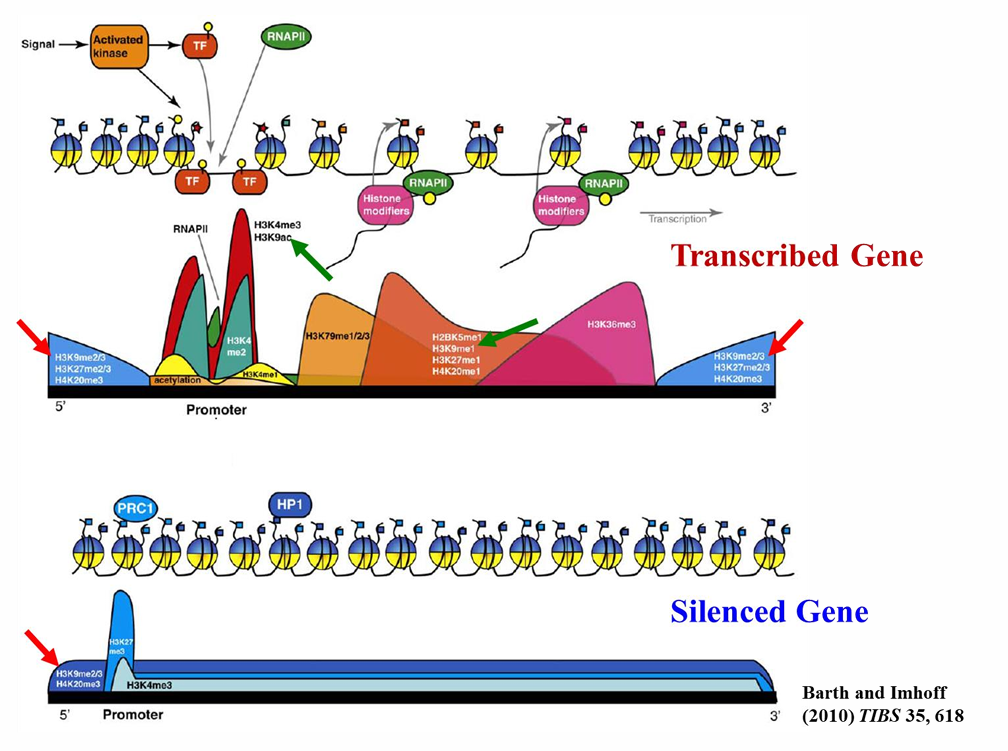
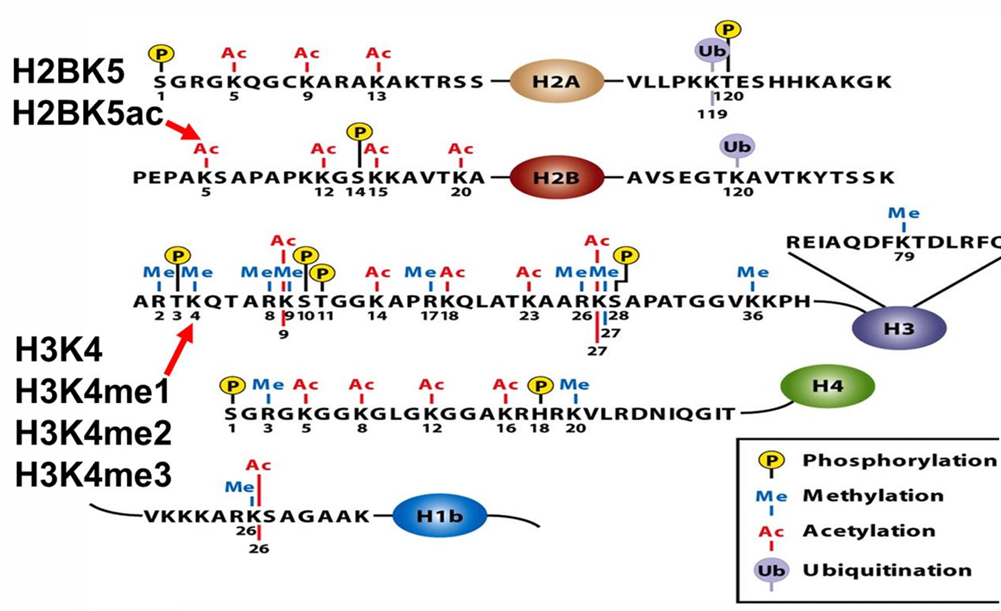
histone code
trimethyl lysine 27 on histone 3 = H3 K 27 me3
DNA transcription regulated by posttranslational modifications to histones
to designate a modification, (histone protein) (amino acid letter) (# position of aa at N terminus) (covalent modification)
DNMT
DNA methyltransferase
hypermethylates CpG island to silence promoter
CpG island
hypermethylation silences promoter, hypo activates it
cluster of CG bonds that can be methylated
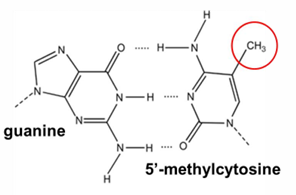
methyl group of 5methyl cytosine protrudes into major groove
affects binding transcription
DNA methylation
NOT histone methylation,
CG, C can be methylated > CpG
euk transcription requires positive regulation
default is off, must be activated
activators, architectural regulators, chromatin mod, remodeling proteins, coactivators, general transcription factors
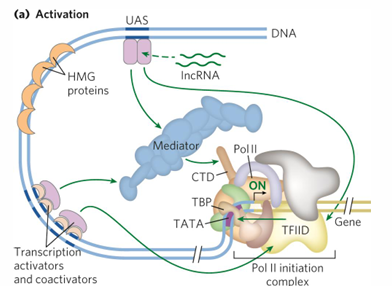
architectural regulators
help DNA looping, bind between promoter and activator/repressor
abundant in chromatin
HMG = high mobility group, help remodeling and activation
high mobility group
help chromatin remodeling and transcription activation
coactivators
help communication between activators, Pol2, and general transcription factors
repressors may bind in their place
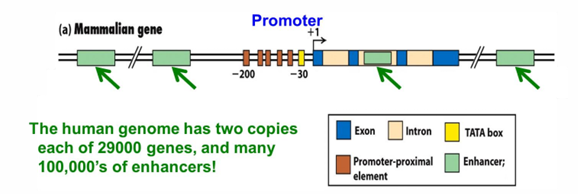
transcription start site +1
has promoter proximal elements upstream -40>-200
enhancers promoter distal positions
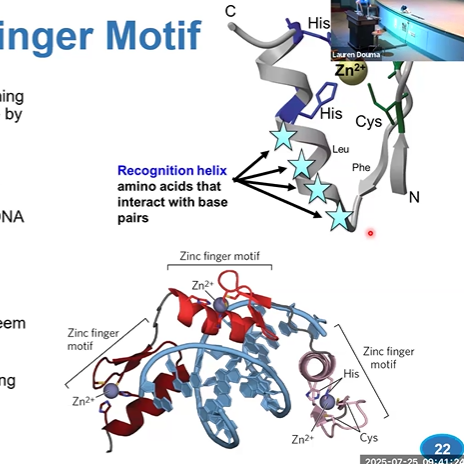
DNA binding domain is made to position a ____ into the ____
recognition helix; major groove
most eukaryotic sequence-specific DNA binding proteins have one of these motifs:
helix-turn-helix, homeodomain
classical zinc finger, nuclear receptor zinc finger,
leucine zipper proteins, helix-loop-helix proteins
**all have recognition alpha helix
DNA binding domain
has TF bind site, activation domain, made up by TFs
zinc finger
30 aa long, elongated loop held by Zn2+ ion;
antiparallel B sheets, 2 cysteines, turn, alpha helix, 2 histidines
cysteines and histidines coordinate Zn to make 3D structure
prokaryotic v eukaryotic transcription factors structure
we have monomer with multiple DNA binding sites,
prokaryotes have to have dimers
the more recognition helixes,
the more specific
leucine zipper
2 roles, bind DNA specifically, and protein-protein interactions to bring subunits together;
zipper region has lots of zipper, interacts with Hphobic on other helix
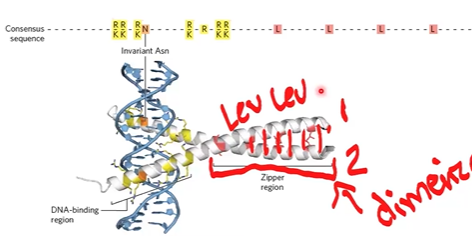
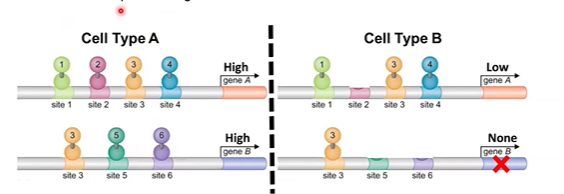
combinatorial control
mixing and matching protein family variants
activation domain
interact with other proteins to recruit them to gene and build the complex for transcription;
coactivators, HAT, CRC (chromatin remodeling complex), mediator, PIC (preinitiation complex);
area of protein-protein interaction
chromatin remodeling
HATs do covalent modifications, acetylate histone tails
CRC use energy to push nucleosomes
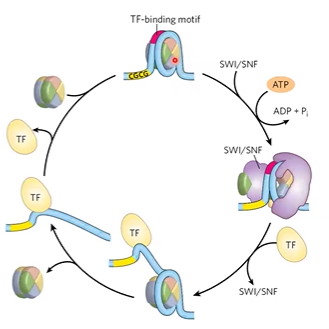
SWI/SNF
a CRC that binds to nucleosomes, removes histone proteins, opens to recruit other proteins
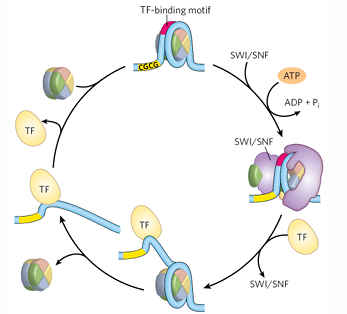
pioneering transcription factor
can access DNA as heterochromatin (only one, very rare);
opens it through activation domain, recruits HATs and CRCs
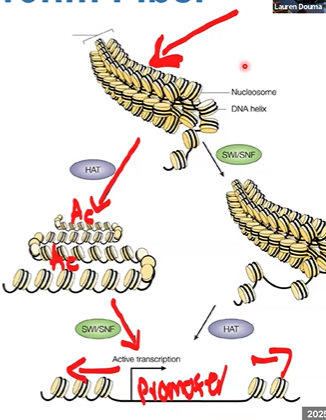
eukaryotic transcription initiation
activators (TFs) bind to regulatory sites
activation domain recruits coactivators: HATs, CRCs, TFs
recruits mediator, bridges activators and RNA polymerase at promoter
recruit RNA poly to recruiter
promoter proximal sites
near promoter
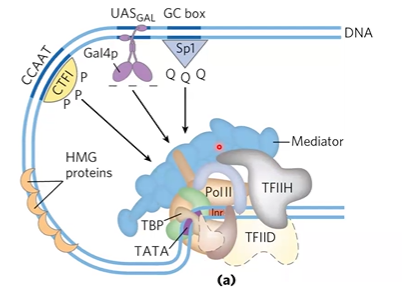
mediator
acts as bridge between activators and RNA pol 2 at promoter;
large complex, major eukaryotic coactivator (doesnt bind to DNA)
binds different activators with 30 polypeptides, binds to CTD
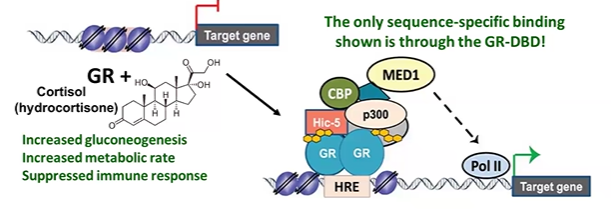
GR nuclear hormone receptor example
GR binds to HRE at promoter proximal sites or enhancers
GR activator domain recruits coactivator with HAT activity
recruits mediator (MED1), which interacts with PIC at promoter.
eukaryotic RNA polymerase
RNA poly 2, makes mRNA and ncRNA (c term tail)
RNA poly 1 makes rRNA
RNA poly 3 makes tRNA and ncRNAs
similar to prokaryotic core enzyme
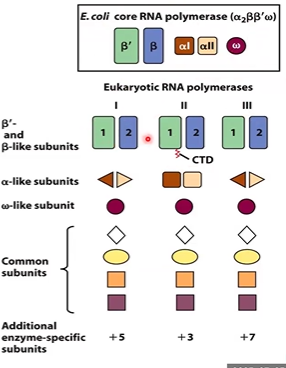
core enzyme v holoenzyme in prokaryotes
holoenzyme has sigma, which recognizes promoter sequences
why do we have to bring RNA polymerases to the promoter in eukaryotes?
it can’t find the promoter by itself (eukaryotic lacks sigma)
general TFs
help gets pol 2 to the promoter
TFII___
required at every rna poly 2, protein-protein
TFs
proteins that help pol 2 form active transcription complex
TF2D
read and binds to promoters
made of TATABP and 13 TAFs
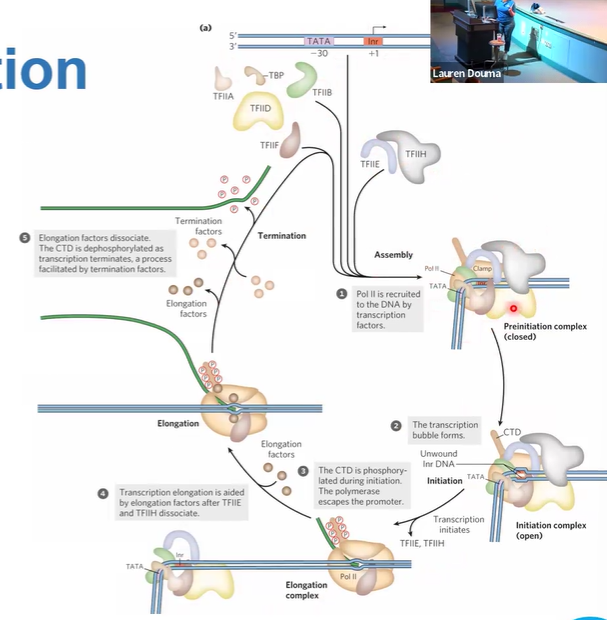
TATABP
locates and binds TATA
TAFs = TBP-associated factors,
transcription initiation & locating promoters
DABTFPEH
TF2D finds it
recruits TF2A maybe
recruits TF2B which binds to DNA and TBP,
TF2F & RNA pol 2 binds TF2B
TF2E and TF2H enter
mediator gets everything together
TF2H Plates cystines of CTD of pol 2
elongation, TF2E and TF2H leave
leaves promoter, transcribes until terminator
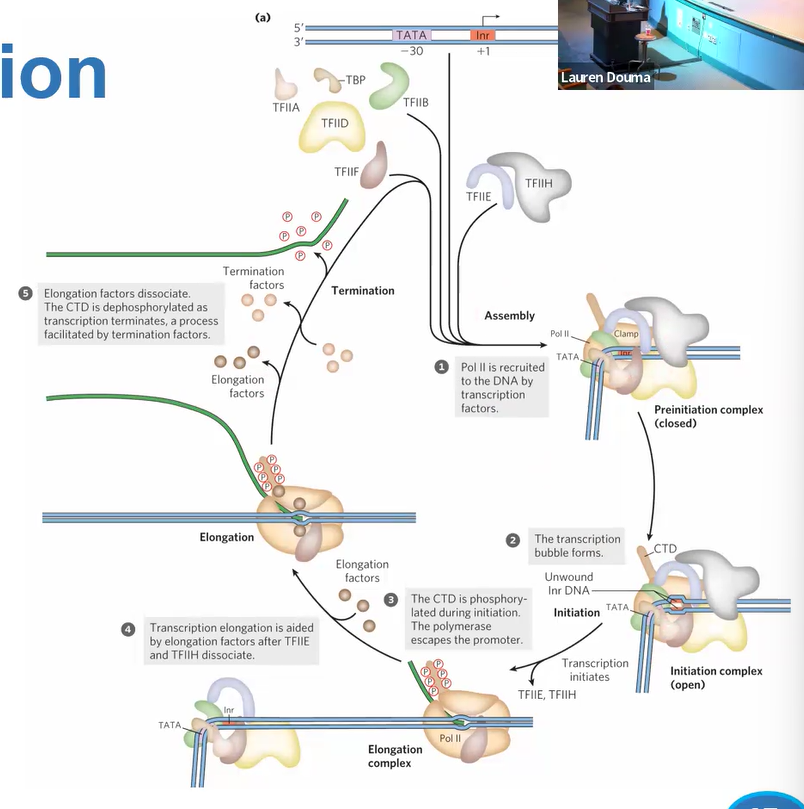
TF2F
binds RNA pol2 to TF2B
TF2H
has helicase (needs ATP) and kinase;
Plates pol 2 at CTD
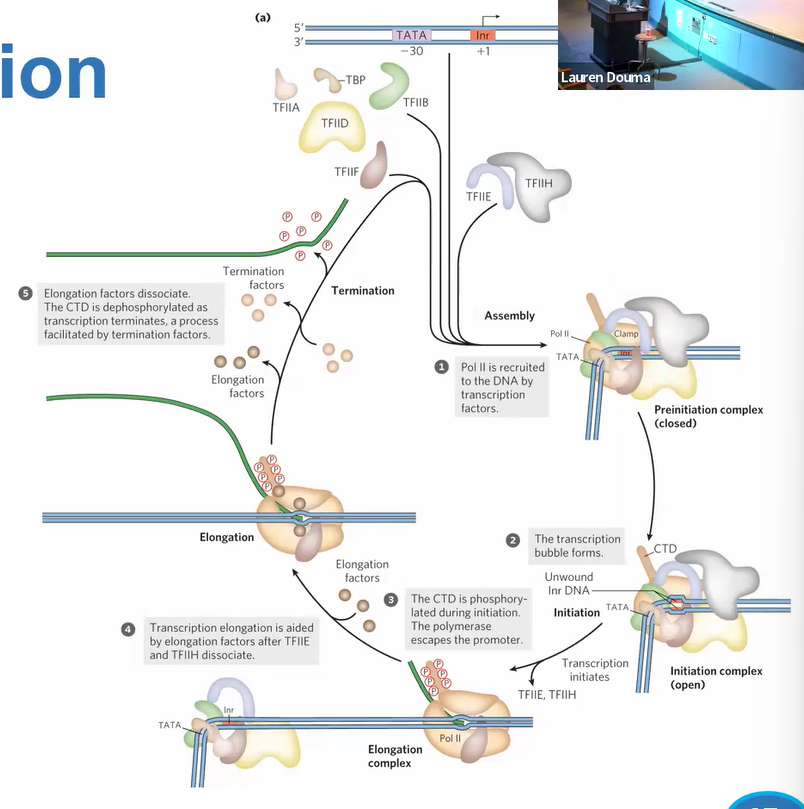
when does transcription start for real
mediator bridges activators and pol 2
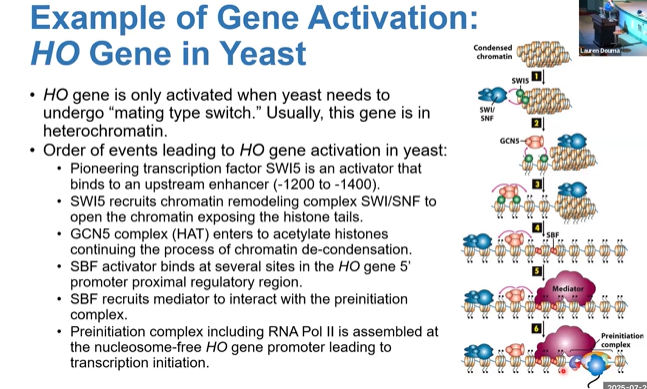
example of gene activation in yeast
starts as heterochromatin, is activated to beads on a string
negative gene regulation
competitive binding, corepressor, altering assembly of PIC, provide dock site for HDAC
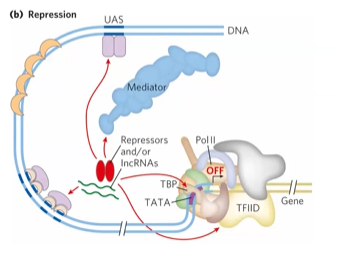
corepressor
prevents interaction with mediator
HDAC
condenses chromatin,
HDAC deacetylates lysine, restoring positive charge
easily reversed
heterochromatin
HMTs (histone methyl transferase) help condense with chromatin associated proteins
HP1 = docking site for HMTs
RNA pol 2
can initiate de novo transcription at a promoter in the absence of a primer
is incapable of locating a promoter by itself
synthesizes RNA from genes located in euchromatin
has a similar subunit structure to prokaryotic core enzyme
enhancers
promoter distal elements (but can be up down or in coding sequence)
cis-acting elements
near the gene they act on