Week 9: DNA Polymerase + Replication
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
fractionation
activity assay to find which fraction has DNA polymerase in it
grow E.coli in large amounts w/ protein of interest
break open cells
prepare soluble extract
fractionate/separate extract to resolve different proteins → repeat
can do based on size, solubility, etc.
confirm activity via activity assay
look for incorporation of radioactivity in polymerized DNA
polymerized DNA has different solubility than free NTs
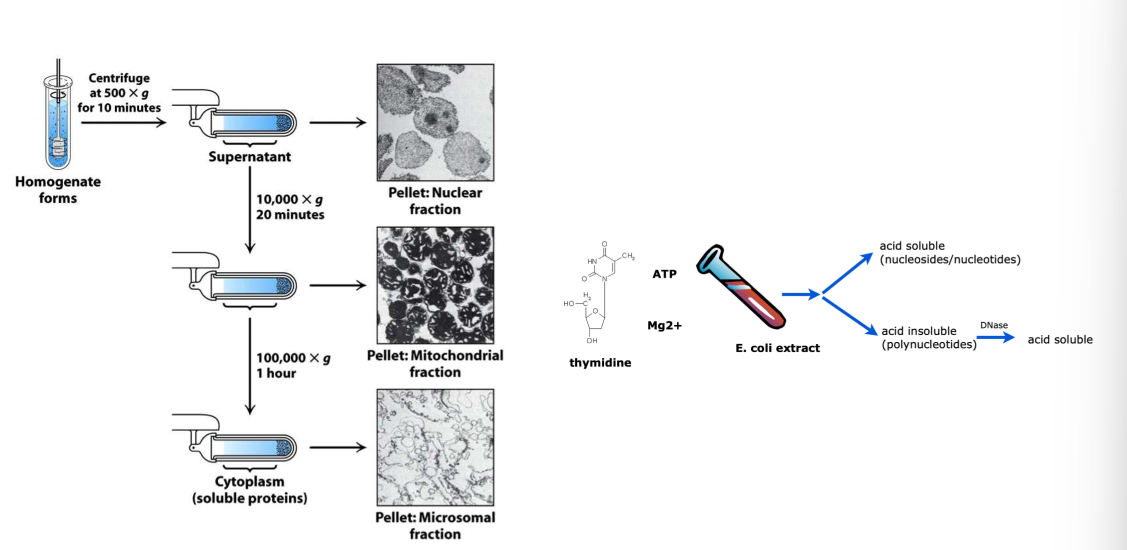
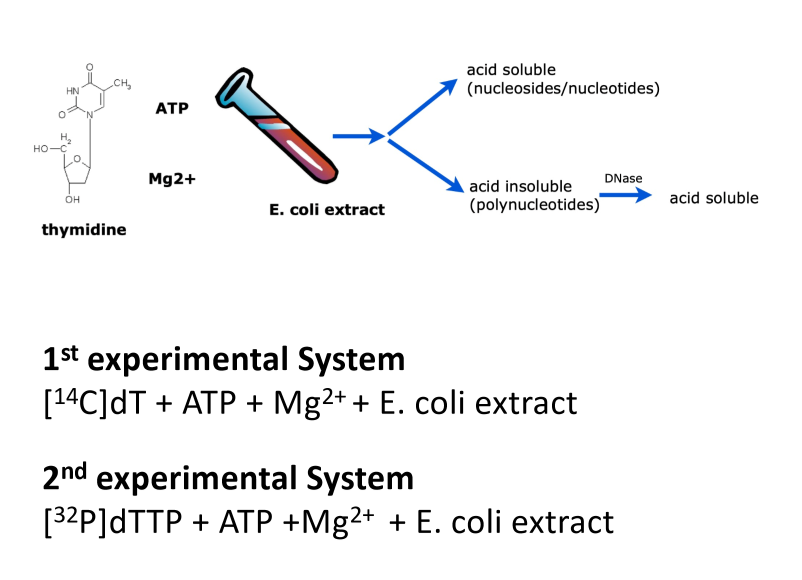
Kornberg experiment
experiments that demonstrated enzymatic synthesis of DNA + identified DNA polymerase as enzyme responsible for DNA replication → DNA Pol I
mixed radio-labeled dT or dTTP w/ isolated fraction
added acid to reaction + separated into 2 fraction via centrifugation:
acid soluble
acid insoluble
acid insoluble = could be polymerized DNA → polymerase is present in mixture
verified acid insoluble pellet contained polymerized DNA by treat w/ Dnase
Dnase specific to DNA → cleaves phosphodiester backbone → all NTs free
real nucleotide polymers broken up → soluble again in acid conditions
radioactive moves from pellet into soluble fraction
NTs could be stuck in middle of protein aggregations/interact w/ protein
need to discern b/w free stuck NTs or free NTs from DNA
DeLucia + Cairns
isolated mutant E.coli w/ no DNA pol I activity
DNA pol I = prototype for all DNA polymerase
enzymatically + structurally
Nicholas Kornberg
discovered DNA pol I through series of fractionation experiments
Tom Kornberg
discovered DNA pol III
polymerase responsible for replicating E.coli chromosome in vivo
DNA polymerase I
involved in cleanup during replication/recombination/repair
abundant but insufficient for replication of E.coli chromosome
rate = 600 NT/min
slower than observed for replication fork movement
low processivity → falls off easily
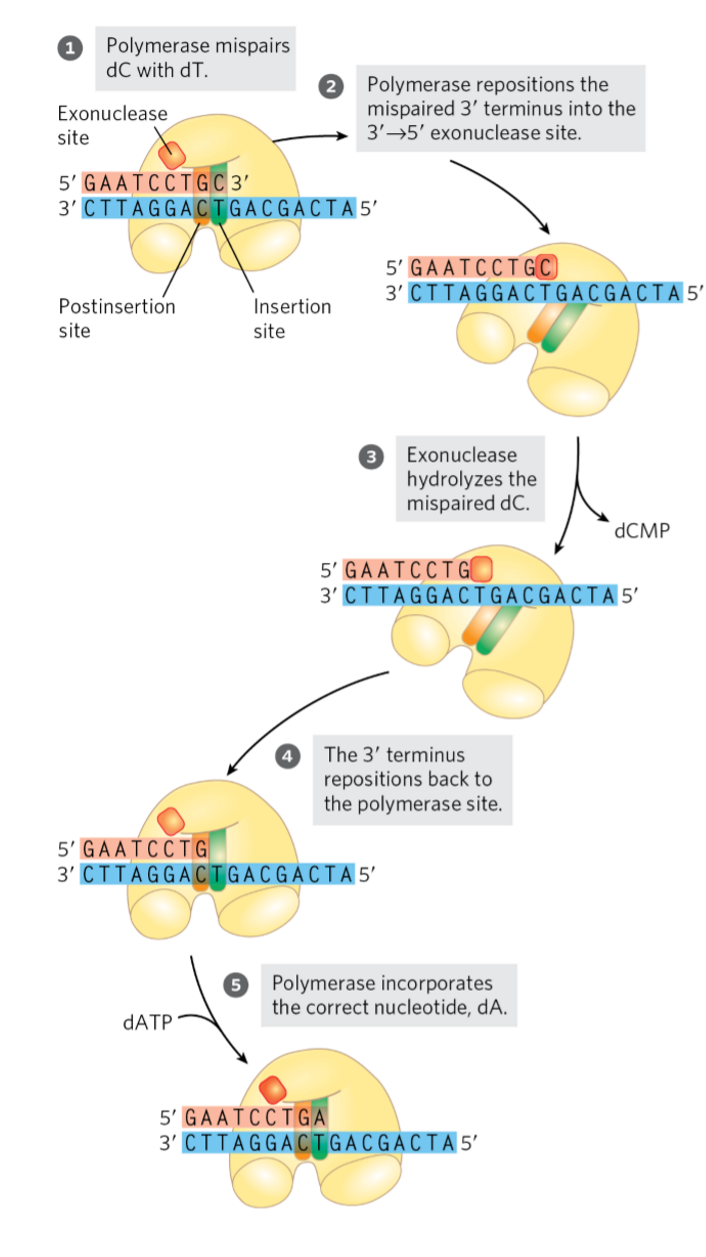
3’→5’ exonuclease activity = proofreading
5’→3’ exonuclease = nick translation
removes RNA/DNA hybrids made by primase
remove RNA primer + add DNA behind
DNA polymerase II
involved in DNA repair
slow polymerization rate
intermediate processivity
3’→5’ exonuclease activity = proofreading
DNA polymerase III
principal replication enzyme in E.coli
3’→5’ exonuclease activity = proofreading
fast polymerization rate
high processivity
DNA polymerase IV + V
involved in specialized DNA repair
no exonuclease activity in 3’→5’ or 5’→3’ direction
very low polymerization rate = 1-3 NT/s
very low processivity
nick translation
break/nick in DNA phosphodiester backbone moved along w/ enzyme
5’ → 3’ exonuclease activity
carried out by larger or Klenow fragment in DNA pol I
makes nicks + removes nucleotides as enzyme moves in 5’→3’ direction
normal polymerase active site adds NTs onto 3’OH end of strand
important in
DNA repair
removal of RNA primers during replication
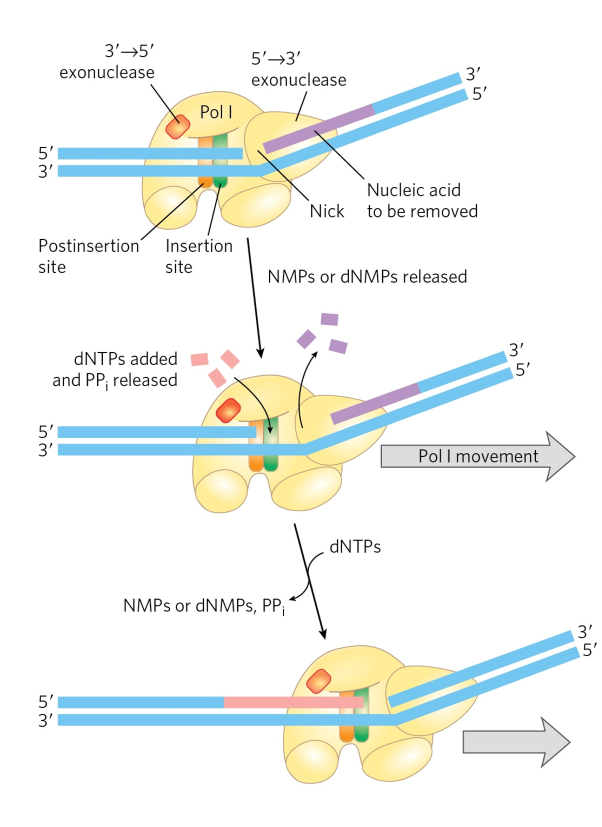
3 to 5 exonuclease
proofreading function
polymerase reads “backwards”
DNA synthesized 5’ → 3’ direction
wrong base added
read backwards along sequence to remove wrong base
5 to 3 exonuclease
nick translation function
aka large fragment or Klenow fragment
domain in front of enzyme + performs nick translation
only in DNA pol I
mild protease treatment separates domain from enzyme
DNA polymerase
synthesizes new DNA polymers in template driven process in 5’→3’ direction
requires:
single-stranded template
deoxyribonucleotides w/ 5’ triphosphate = dNTPs
Mg2+ ions = co-factor for polymerase
aspartic acid residues = (-)
Mg2+ has to be in right spot to stabilize transition state at 3’OH
catalyzes reaction
annealed primer w/ free 3’OH
often RNA
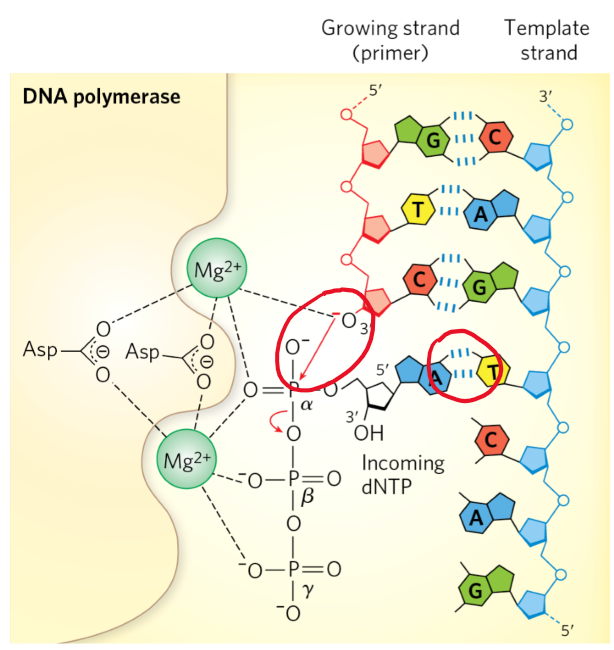
DNA polymerase reaction mechanism
primer already bound to DNA template strand
incoming dNTPs form WC bonds w/ template strand
coordination w/ Mg2+ stabilizes 3’ OH + a-phosphate
new bond forms b/w a-phosphate of incoming NT + 3’OH of primer
beta + gamma phosphate of NT released as PPi new phosphodiester bond forms
as long as template strand is available → new substrate generated for next round of polymerization
once last NT reached → stops
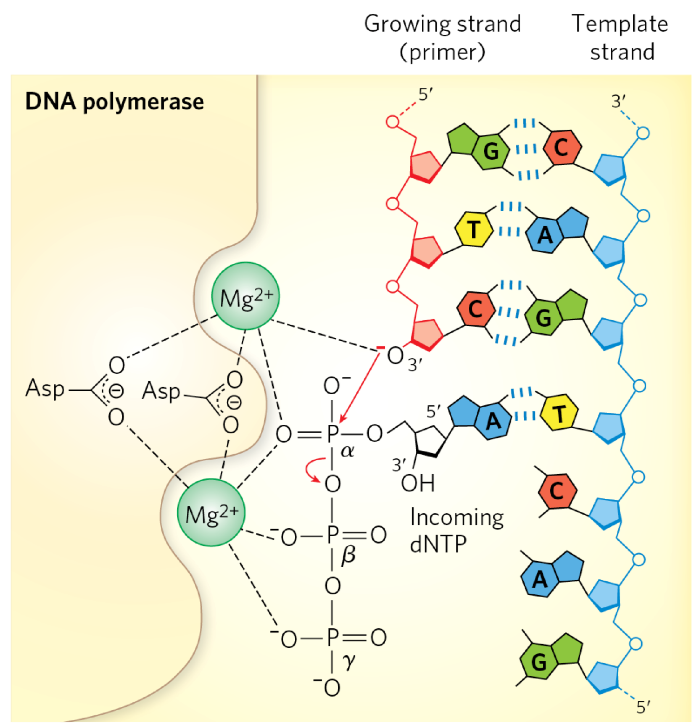
DNA polymerase template
guides polymerization according to WC base-pairing rules
needs to be single-stranded
double-strand → all H-bonds satisfied = can’t add more NTs in
need helicase to unwind
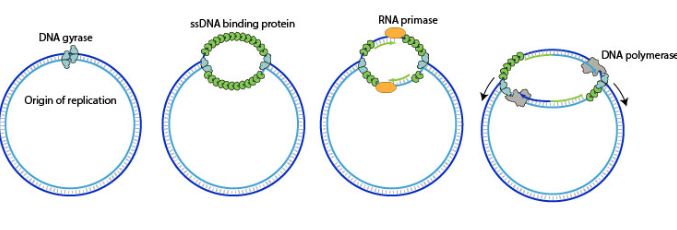
DNA helicase
unwinds parental DNA to make single-stranded template
in vitro assay
radio-label 1 strand of dsDNA fragment
add purified enzyme in gradient = ⬆ [enzyme]
look for ability of protein to unwind DNA via ssDNA product formation
DNA segments become smaller as ⬆ [enzyme]
single-stranded binding protein (SSBP)
binds to single-stranded DNA to prevent spontaneous re-annealing + intra-strand interactions
in E.coli = SSP
in humans = RPA
sequence-independent = doesn’t make specific contact w/ DNA
needs to be jiggly = allow polymerase to read DNA
shape discrimination
contributes to fidelity of DNA replication via base-pair geometry
active site flexible enough for catalysis of properly matched bases
incorrect BP = binding site contorts = less favourable
prevents replication of wrong BPs
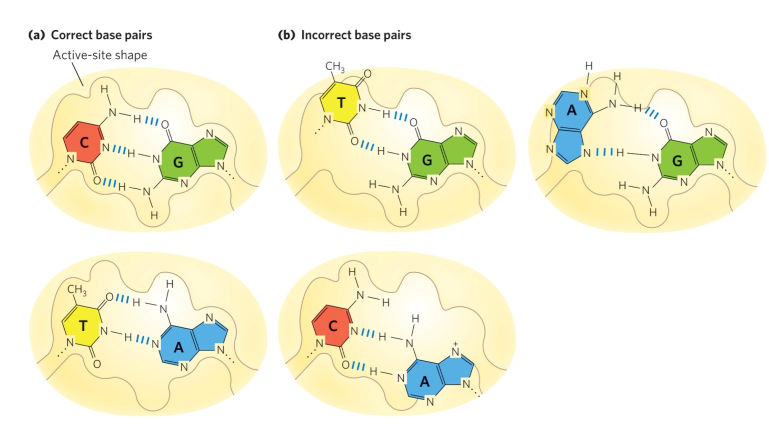
DNA polymerase error correction (proofreading)
translocation of enzyme inhibited when wrong NT added
3’→5’ exonuclease activity → removes newly added NT
domain flips to active site
mismatch fits into space for mismatched bases
favours non-Cannonical BPs
exonuclease cleaves off NT
semi-conservative replication
parent + daughter strands wind together
daughter strands different from e/o but net product w/ parent = identical copies
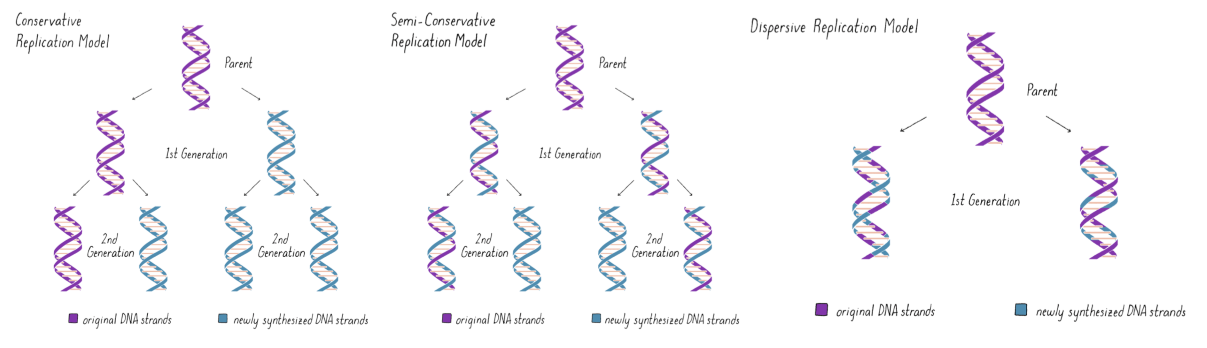
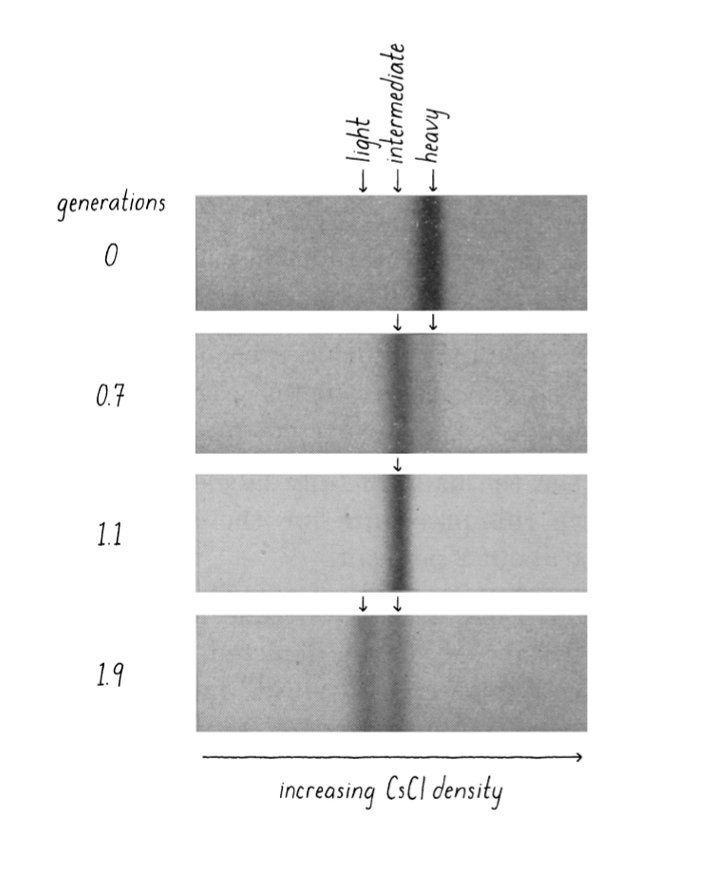
Meselson-Stahl experiment
grow E.coli in “heavy” nitrogen environment to label DNA
bacteria copy entire DNA complement (genome) before every cell division
DNA = heavy
moved into normal N-containing medium
cell divide once more
DNA = lighter
separate DNA by density via cesium chloride density gradient
heavy → travels more
results support semiconservative replication
after 1 generation = DNA intermediate
conservative replication ❌
after 2 generation = DNA either all light or intermediate
semiconservative replication ✅
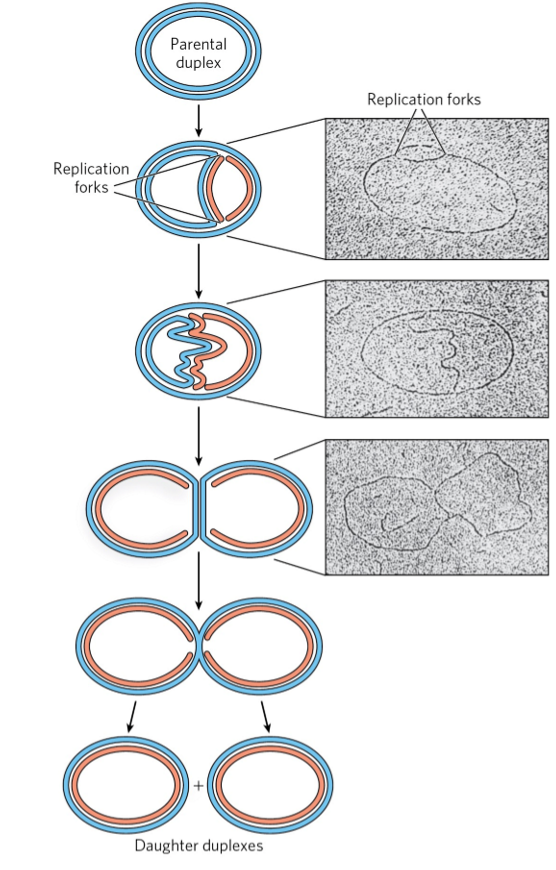
replication fork
dynamic points where parent DNA is unwound + separated strands are replicated
both strands replicated simultaneously
both ends of bacterial chromosome have active replication forks → bidirectional replication
denaturation mapping
selective denaturing of sequences rich in A-T base pairs
AT = only 2 H-bonds + ⬇ base stacking = easier to melt (separate)
provide landmarks along DNA molecule
generates reproducible pattern of single-stranded bubbles
origin
location where replication loops are initiated
bidirectional replication
process where DNA replication occurs simultaneously in both directions from single origin of replication, resulting in two active replication forks
semi-discontinuous replication
mode of DNA replication where 1 strand (leading strand) is synthesized continuously while other strand (lagging strand) is synthesized in short segments (Okazaki fragments) due to the antiparallel nature of the DNA double helix
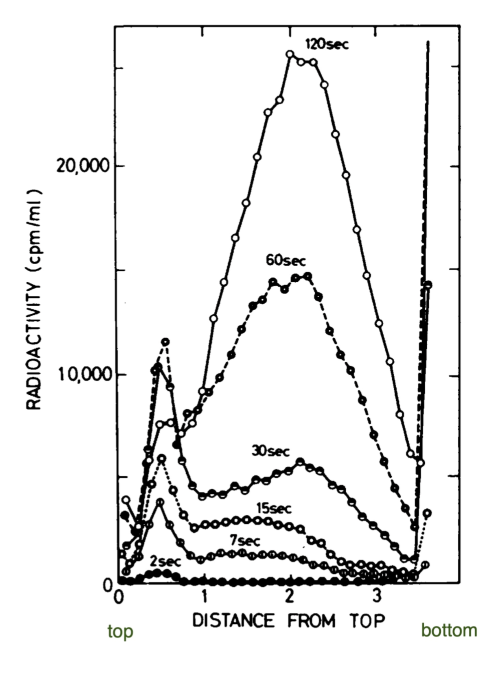
Okazaki’s experiment
method
pulsed E.coli w/ radioactive thymidine
newly synthesized DNA labeled = “hot'“
used alkaline sucrose density gradient
separate DNA based on size
assay fractions from centrifugation experiment at various time points
DNA synthesis stops once cells are lyzed
can see “snap” shots of reaction
results
trace = time point
dot = point along gradient density
further down gradient = larger fragment w/ “hot” Thymidine
over time → see longer DNA fragments + still see short ones
supports discontinuous replication model
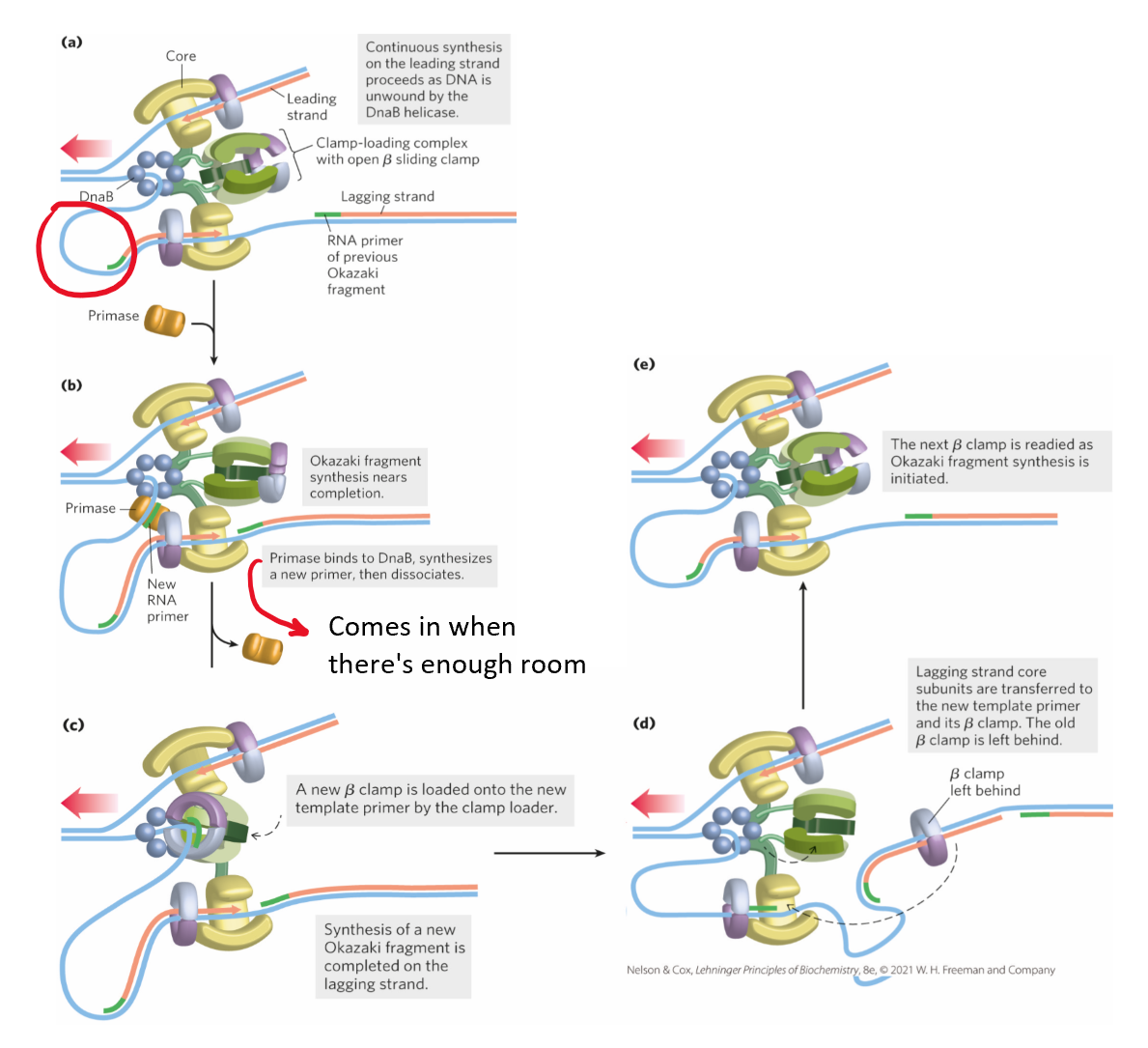
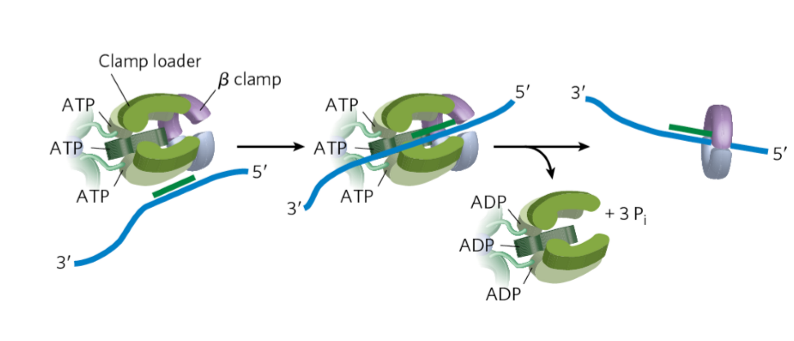
DNA polymerase III clamp loader
loads clamp onto DNA-RNA hybrid
requires ATP binding + hydrolysis
ATP physically opens up ring
ring snaps back around region
helps w/ processivity of polymerase
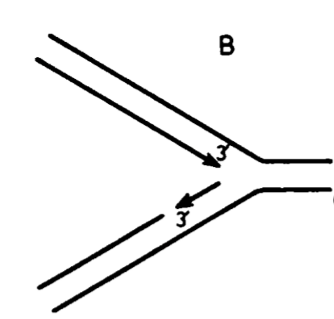
leading + lagging strand synthesis
helicase (DnaB) unwinds DNA at replication fork
single-stranded lagging strand coated by SSBP
leading strand synthesized in one piece
lagging strand synthesized in fragments = Okazaki fragments
primase = adds primer to make free 3’OH for polymerase
clamp loader loads on clamp
ensures synthesis is processive
lagging strand synthesized until primer on older Okazaki fragment reached
clamp loader reloads clamps on new primer/template RNA-DNA hybrid until synthesis completes
DNA pol I removes RNA primer + replaces w/ DNA via nick translation (5’→3’ exonuclease)
DNA ligase seals remaining nick in phosphodiester backbone