1.4 Mobile Genetic Elements and genome fluidity
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
59 Terms
Mobile genetic elements (MGEs)
are DNA segments that can move within and between genomes, contributing to genetic diversity and adaptability in organisms.
Do all isolates have the same MGE gene content
No, isolates can have different sets of MGEs, leading to variations in their genetic makeup and adaptability.
genomic island (GI)
a part of a chromosome that has evidence of being acquired via
horizontal gene transfer.
What would be evidence that a part of a chromosome is a genomic island
Variably present in strains
Have phage integrases and plasmid conjugation systems
mostly inserted at tRNA genes
Why are genomic islands mostly inserted at tRNA genes
these regions provide a stable and conserved integration site for foreign DNA
Wats an example of evidence that a part of a chromosome is a GI
Difference GC%
What types of genes are usually encoded in GIs
often carry genes offering a selective advantage
for host bacteria to overcome the fitness cost
of carrying the island.
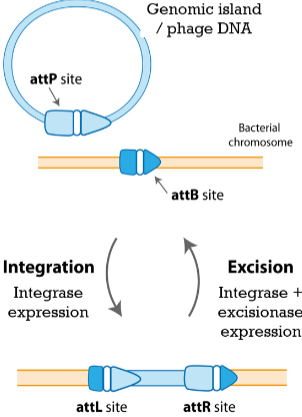
Integrases
sequence-specific recombinases that catalyze insertion gy recombination
Can integrases participate in excision
Yes, but require a second
protein (excisionase)
Describe integrase recombination
between 15- to 20-bp motifs on the GI / phage (attP) and
bacterial chromosome (attB). Results in the generation of two hybrid sequences (attL and attR)
What is the effect of a modular design on the diversity of bacteriophages (phages)?
Modular design allows for the exchange of genetic modules between different phages through genetic recombination.
Greater diversity
do you have any
thoughts on why the integrase
(int) gene is almost always
situated at one end of an
integrated GI / prophage?
The integrase enzyme, encoded by the int gene, mediates site-specific recombination between the phage/ GI DNA and the bacterial chromosome. Positioning the int gene at one end ensures efficient access to the integration sites in the host genome
Why does integrase mediated integration not lead to mutation
When integration occurs, the chromosomal tRNA gene loses its original 3’ end but gains the 3’ end provided by the phage or genomic island so tRNA function isn’t disrupted
Integrative Conjugative Elements (ICEs)
Genomic islands that can excise from their chromosomal location, replicate, conjugate
themselves to another cell, and reintegrate into the new host’s chromosome
Are all genomic islands mobile?
Some genomic islands are non-mobile (e.g., due to mutation of the conjugation genes)
Why is there so much variation in GIs between strains
Genomic islands are recombination hotspots (could be due to low GC%, repeat sequences,
homology amongst common function genes, etc.)
SOS response
A cellular response to DNA damage that induces the expression of DNA repair genes and can lead to mutagenesis.
LexA
A repressor protein that regulates the SOS response by inhibiting the expression of repair genes until DNA damage occurs.
recA
A protein essential for homologous recombination and the SOS response, facilitating the repair of DNA damage by promoting the cleavage of LexA.
What does SOS response have to do with DNA excision and transfer of mobile gene elements
SOS response activates RecA which deactivates LexA repressor proteins, allowing the excision and transfer of mobile genetic elements
How do most mobile genomic islands transfer from one cell to another
conjugation
If a species like S. aureus don’t have conjugation genes, then how can they transfer mobile gene elements
they are packaged into a phage particle,
released from the cell, and undergo a phage-type infection
before integrating into the new host chromosome
SaPI transfer
is a mechanism in which Staphylococcus aureus pathogenicity islands are packaged into phage particles and transferred to other bacteria through infection, facilitating horizontal gene transfer.
The mechanism of SaPI transfer
A “helper phage” is induced by the SOS response, excising
out of the chromosome, replicating, and making
phage particle proteins (right). The SaPI also
excises out of the chromosome and replicates,
with SaPI genomic DNA being packed into phage
particles that end up with smaller heads.
What is the relationship between the SOS response and
horizontal gene transfer?
the transcriptional repressor proteins of phage and
genomic islands are cleaved in the presence of RecA,
ultimately promoting their induction and transfer
Transposons
elements that insert into other DNA molecules (e.g., chromosome, plasmids) and hence are a common source of mutation
Simple transposons (a.k.a. insertion sequences [IS])
Only contain genes for a transposase, inverted repeats for transposase recognition, and only need a few based of homology for insertion
Complex transposons
Include insertions sequences on both ends and additional genes between them that could have toxin production or resistance
Complex transposons are exploited ____
to make libraries of
mutant strains of a given microbe
How are transposons transferred
horizontally transferred by hitching a ride on a plasmid or genomic
island. Others encode the proteins needed to conjugate
conjugative transposons
encode the proteins needed to conjugate
Plasmids
are small, circular DNA molecules that replicate independently of chromosomal DNA and can carry genes that provide advantageous traits, such as antibiotic resistance.
What types of genes are most often found in a plasmid
virulence factors, antibiotic resistance, etc
conjugative plasmids
plasmids encode all the proteins required to
conjugate themselves
Copy level of a plasmid (low, medium, high)
determines how many times its genetic material is duplicated within the bacterial cell, affecting the level of gene expression and the overall impact on the host cell
Why would a plasmid have a low copy number
if the plasmid carries genes that place a burden on the cell
Why would a plasmid have a high copy number
ensures high expression of plasmid-encoded genes, which can be advantageous for producing large amounts of a protein or other products
Do all bacteria have integrated phages
No
Lytic cycle of a phage
A reproductive cycle in which a phage infects a host bacterium, replicates itself, and ultimately causes the host cell to lyse, releasing new phage particles.
Lysogenic cell of a phage
A bacterial cell that contains a prophage, where the phage genome is integrated into the host's chromosome and can be replicated along with the host's DNA.
Phages in pathogens
often encode virulence factors, e.g., Botulinum toxin, Shiga toxin, etc
How can phages influence expression of virulence factors (like toxins) in Enterohemorrhagic Escherichia coli
If antibiotics are used to lyse open bacterial cells, the patient will actually deteriorate because this lysis releases more toxins
Also as the antibiotics damage DNA, SOS is activated and even more phages are produced which means more toxin produced as well
What determines E. coli pathotype
MOBILE GENETIC ELEMENTS
Enterotoxigenic E. coli (ETEC)
the leading bacterial
cause of diarrhea in the developing world, as well as the
most common cause of travelers' diarrhea
Enteropathogenic E. coli (EPEC)
defined as diarrhea-causing E. coli whose virulence mechanism is
unrelated to the excretion of typical E. coli enterotoxins
Enterohemorrhagic E. coli (EHEC)
can cause diarrhea or hemorrhagic colitis in humans.
Hemorrhagic colitis occasionally progresses to HUS.
Enteroinvasive E. coli (EIEC)
cause a syndrome that is identical to Shigellosis,
with profuse diarrhea and high fever
Uropathogenic E. coli (UPEC)
infections account for more than 80% of uncomplicated
urinary tract infections (UTIs)
How much bacteria is lysed in the ocean by phages every day
1/3 then lysed bacteria tend to clump creating large, heavy particles that sink
How do phages affect the marine carbon cycle
by lysing bacteria, as it sinks they are releasing carbon that becomes available for other microorganisms, thus facilitating nutrient cycling and carbon export to the deep ocean.
Why can we find essentially idential phages in widely separated and different environments
large numbers of phage and bacteria in the troposphere and
stratosphere, some of which fall to earth every day
Uropathogenic E. coli (UPEC) infections
account for more than 80% of
uncomplicated urinary tract infections.
Which of the following is one way in which
UPEC differ from commensal E. coli?
UPEC harbor multiple pathogenicity islands while commensal E. coli do not
Information gained from the study of mobile genetic elements
have led to numerous
advances, e.g., phage therapy, CRISPR-Cas, etc
phage therapy.
A therapeutic approach that uses bacteriophages to target and kill specific bacterial pathogens, often used as an alternative or complement to antibiotics.
Enterotoxigenic E. coli (ETEC) mobile gene elements
pENT plasmids that carry genes responsible for heat-labile and heat-stable enterotoxins, contributing to diarrhea.
Enteropathogenic E. coli (EPEC) mobile gene elements
Have LEE pathogenicity islands that carry genes for adherence and invasion of intestinal epithelial cells.
Also pEAF plasmids that enhance virulence by promoting attachment to intestinal cells.
Enterohemorrhagic E. coli (EHEC) mobile gene elements
Has LEE pathogenicity islands that encode Shiga toxin and genes for adherence, leading to severe intestinal disease.
Also Stxo phages that can transfer Shiga toxin genes between bacteria.
Enteroinvasive E. coli (EIEC)) mobile gene elements
Contain invasion plasmid antigens (Ipa) that enable invasion of intestinal epithelial cells, leading to inflammation and diarrhea.
Also black holes which allow for horizontal gene transfer, enhancing pathogenicity.
Uropathogenic E. coli (UPEC) mobile gene elements
Contain 2 pathogenicity islands, LEE and others which carry genes for adherence, invasion, and virulence factors essential for urinary tract infections.