ETC AND ATP SYNTHASE AND OXIDATIVE PHOSPHORYLATION
OIL RIG **
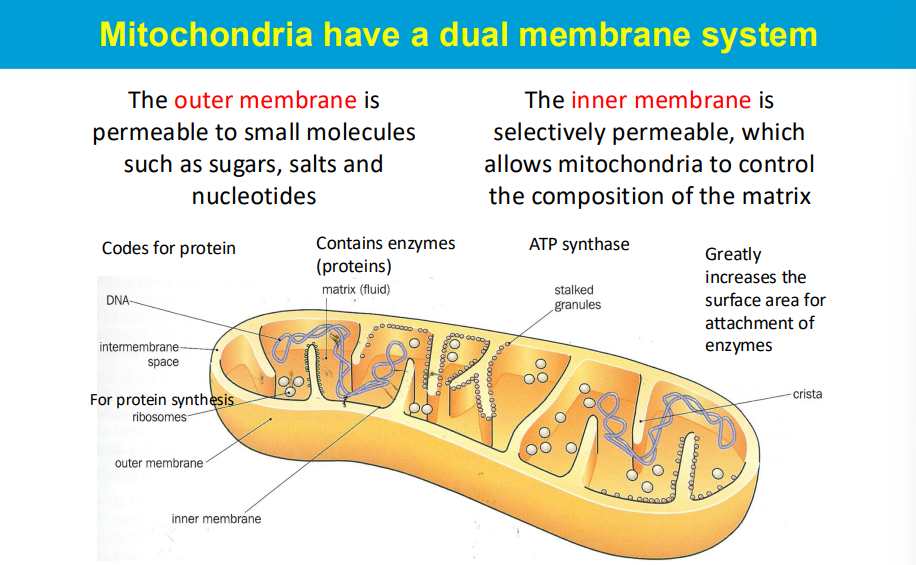
the "matrix" refers to the inner compartment enclosed by the inner mitochondrial membrane, where key metabolic processes like the citric acid cycle occur, while the "intermembrane space" is the narrow region between the inner and outer mitochondrial membranes, acting as a space for protein sorting and signaling pathways
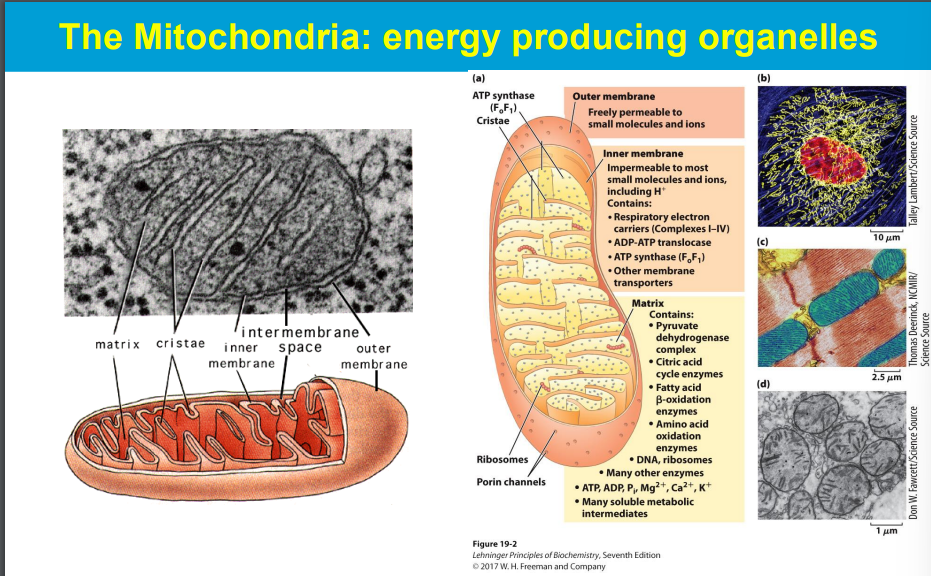
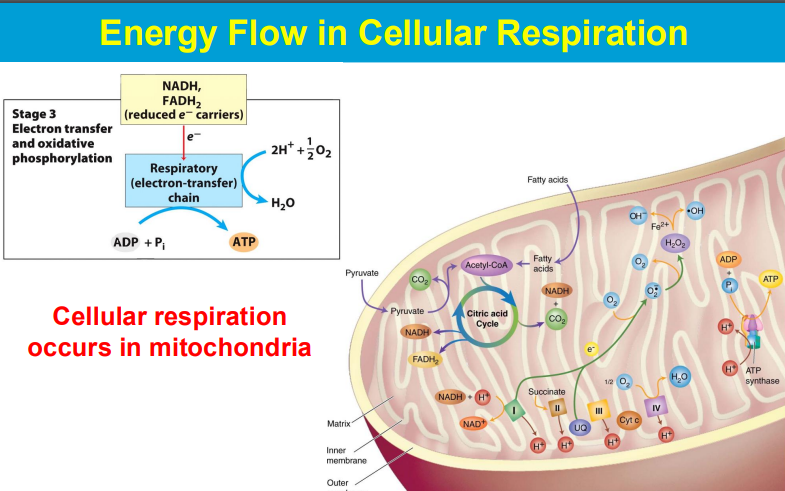
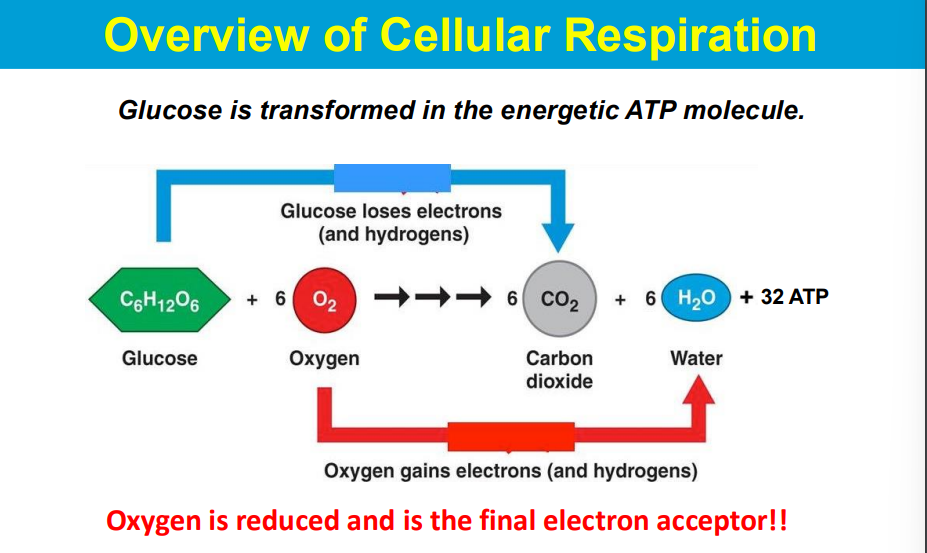
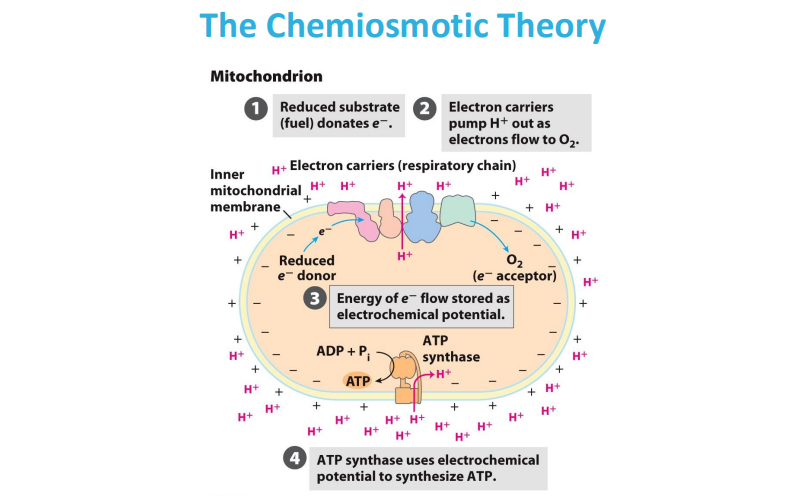
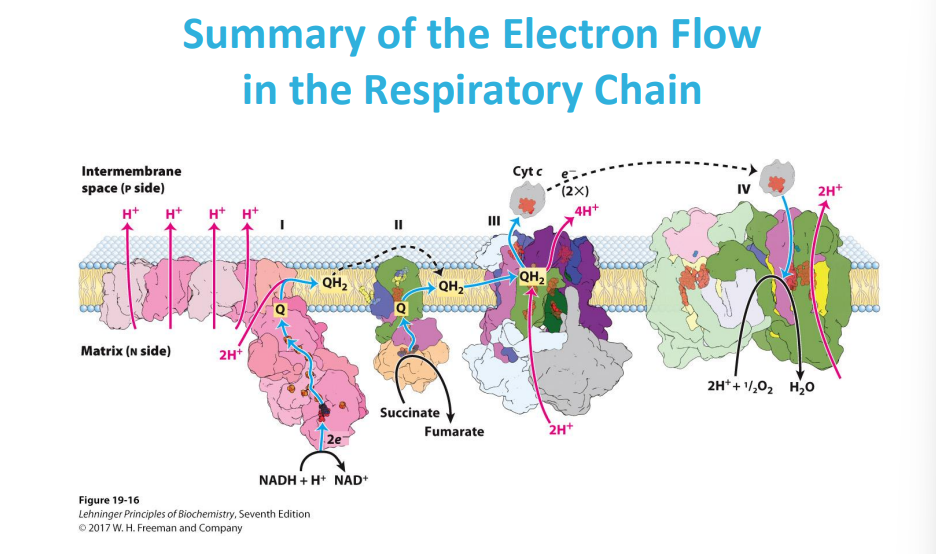
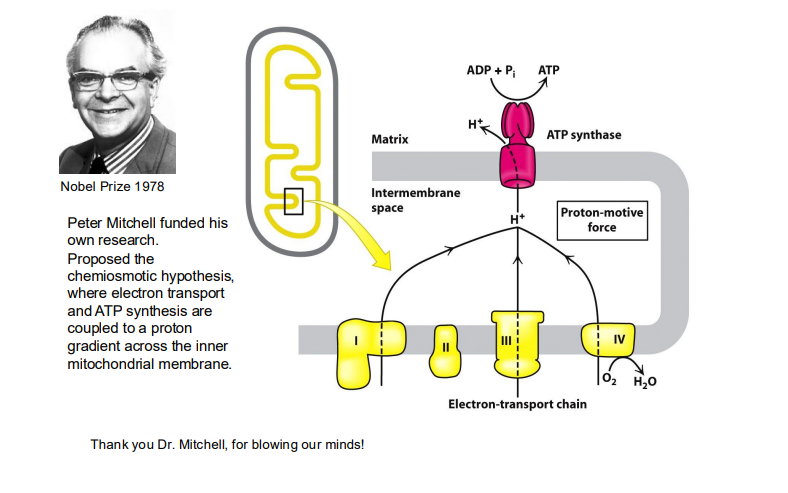
Electrons are passed from NADH and FADH2 (electron carriers) through a series of protein complexes within the mitochondrial membrane (the electron transport chain).
At the end of the chain, Complex IV accepts electrons and transfers them to oxygen molecules, causing oxygen to be reduced to water.
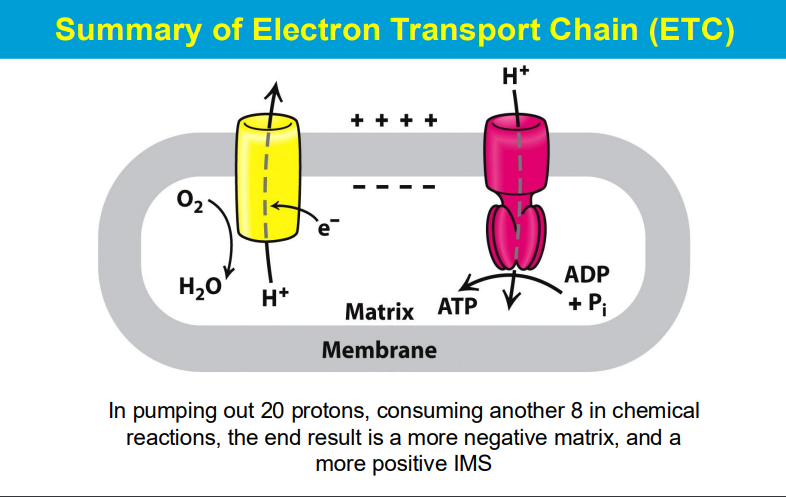
As electrons move through the chain, protons are also pumped across the mitochondrial membrane, creating a proton gradient that drives ATP synthesis.
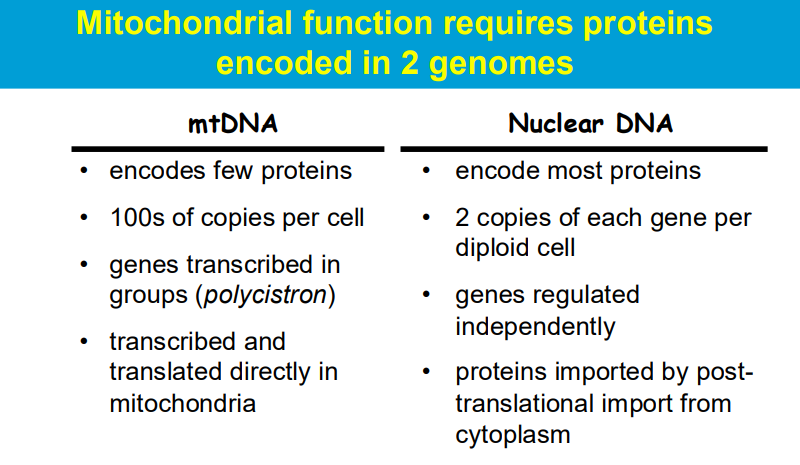
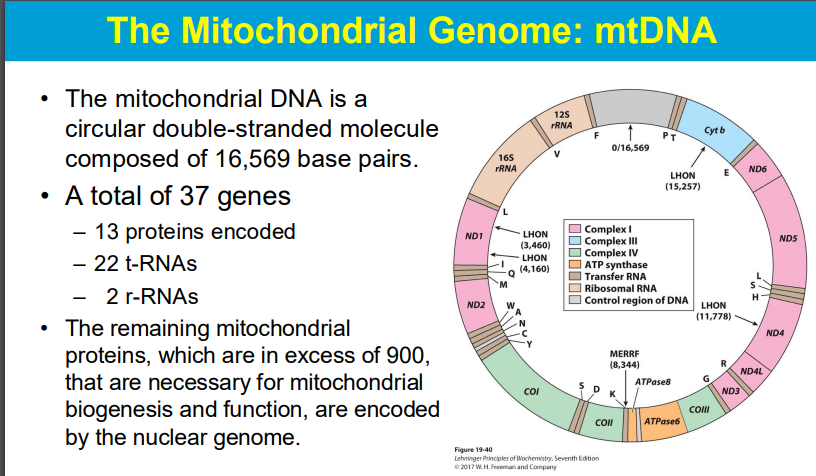
The primary distinction between monocistronic versus polycistronic (mRNA) is that the former generates a specific protein while the latter generates a number of functionally relevant proteins
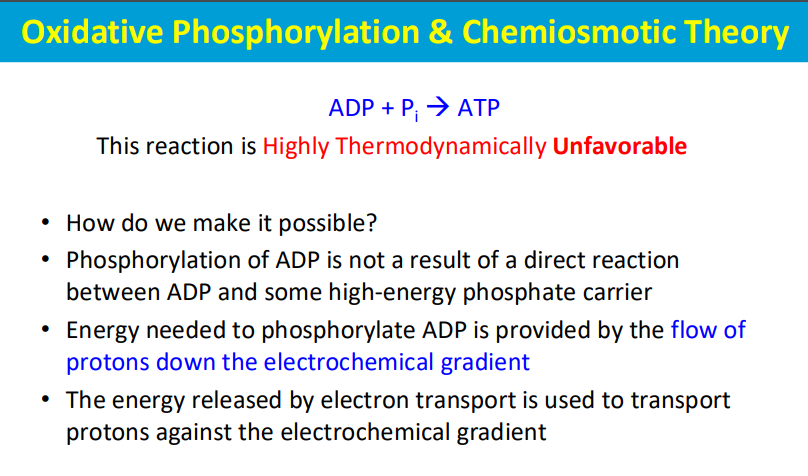
The phosphorylation of ADP (to form ATP) is made possible by coupling it to a highly favorable reaction - the flow of protons down the electrochemical gradient across a membrane, which is powered by energy released during electron transport; essentially, the energy from proton movement through ATP synthase drives the otherwise unfavorable phosphorylation of ADP, making the process thermodynamically viable.
2 different proteins^^
crucial function of specific protein complexes embedded within the mitochondrial inner membrane, which are responsible for harnessing the energy released from the electron flow in the electron transport chain to actively pump protons across the membrane, creating a proton gradient
This protein complex acts as a "proton turbine". As protons flow "downhill" through the complex due to the established gradient, their energy is harnessed to rotate a protein component, which then drives the phosphorylation of ADP to ATP.
** e- donor donates electron to e- acceptor
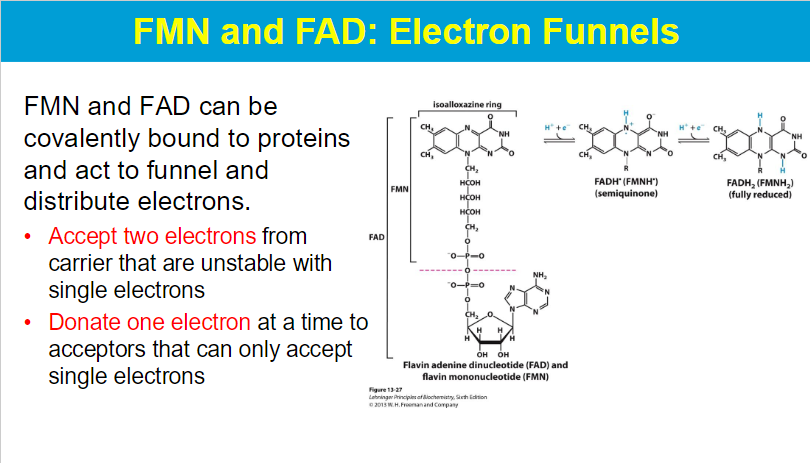
FMN (flavin mononucleotide) and FAD (flavin adenine dinucleotide) are important coenzymes that can be covalently attached to proteins, allowing them to function as electron carriers by accepting two electrons from a donor molecule and then donating them one at a time to different electron acceptors, which are often only able to receive single electrons; essentially acting as a "funnel" to distribute electrons in biochemical pathways.
**electron acceptors of complex I and II
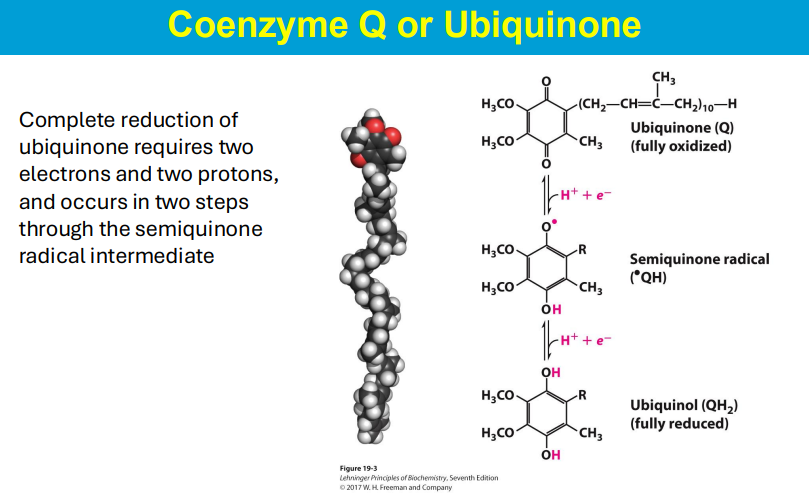
Ubiquinone, also known as Coenzyme Q, is a lipid-soluble molecule within the cell membrane that functions as a mobile electron carrier, accepting electrons from Complexes I and II of the electron transport chain and transferring them to Complex III by converting to its reduced form, ubiquinol, which can freely diffuse across the membrane, effectively transporting electrons with protons from one side of the membrane to the other; essentially acting as a shuttle for electron transfer within the mitochondrial membrane.
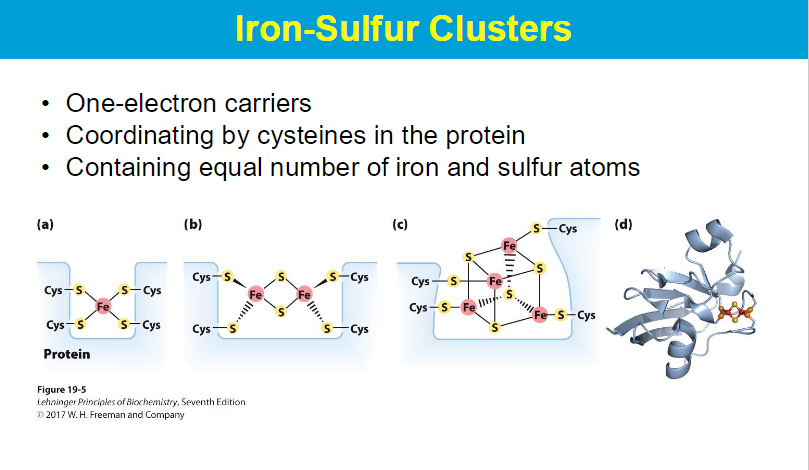
Iron sulfur clusters are coordinated by cysteines through the sulfur atom on the cysteine side chain, which directly binds to the iron ions within the cluster, essentially acting as a ligand that anchors the cluster to the protein structure
Cysteine iron bonds facilitate electron transfer because the sulfur atom in cysteine acts as a readily available electron donor, allowing it to easily transfer electrons to or from the iron ion, which is a redox-active metal capable of changing oxidation states, thus enabling the movement of electrons between different molecules
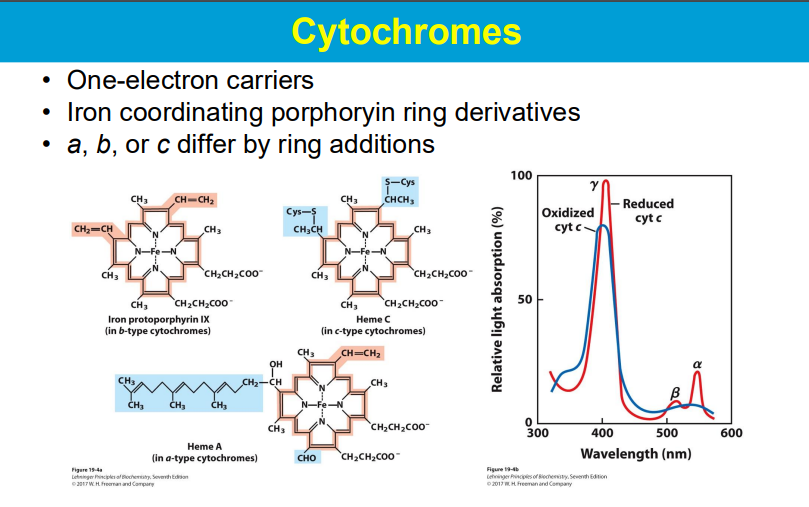
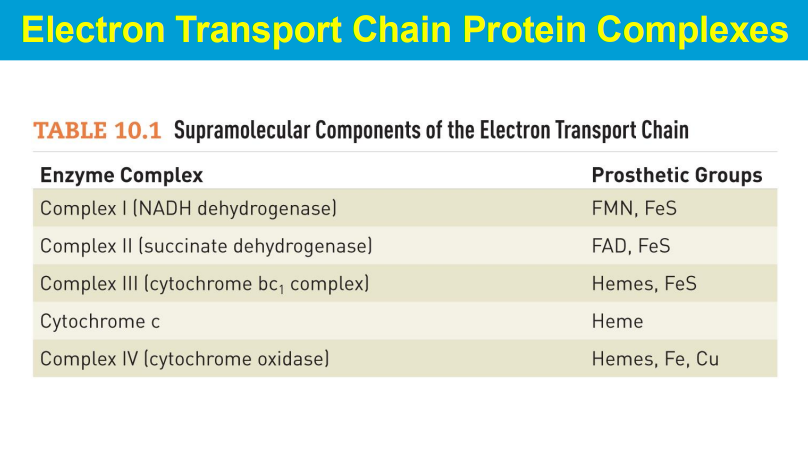
In the electron transport chain (ETC), the energy relationships between complexes lie in the sequential transfer of electrons from higher energy donors to lower energy acceptors, with the energy released at each step being used to pump protons across the mitochondrial membrane, creating a proton gradient that ultimately drives ATP synthesis; this process is known as oxidative phosphorylation.
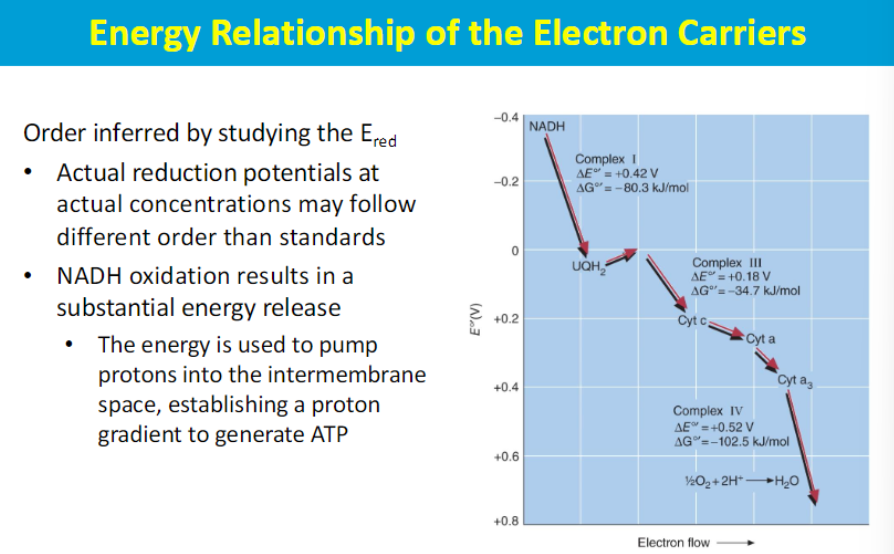
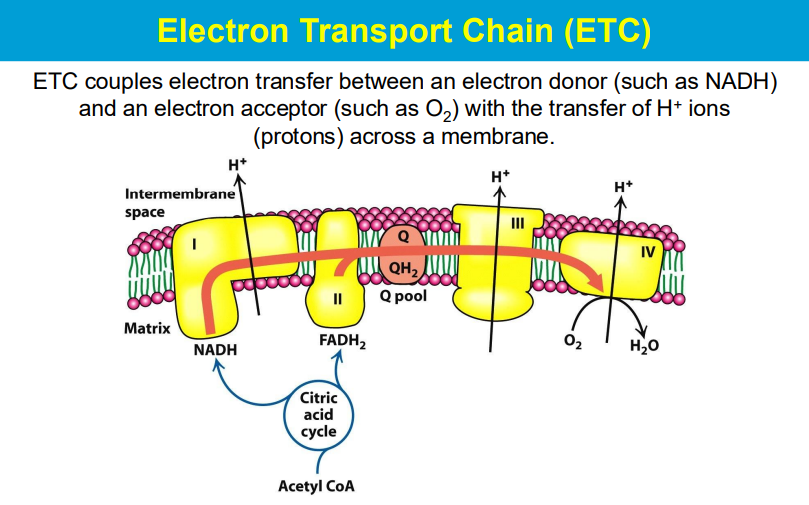
so the electron donor donates electron that goes through the complexes of the ETC with different redox reactions to release energy to pump H+ against the gradient. then the electron is accepted by O2 that is then reduced into H20
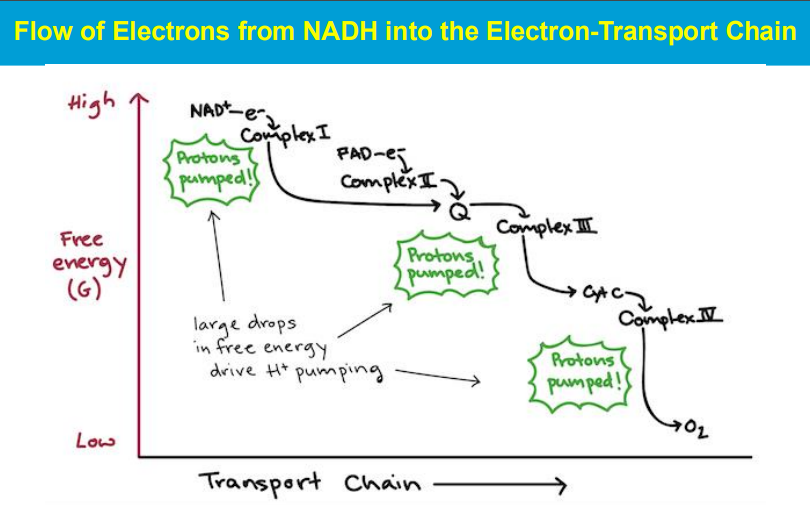
***************
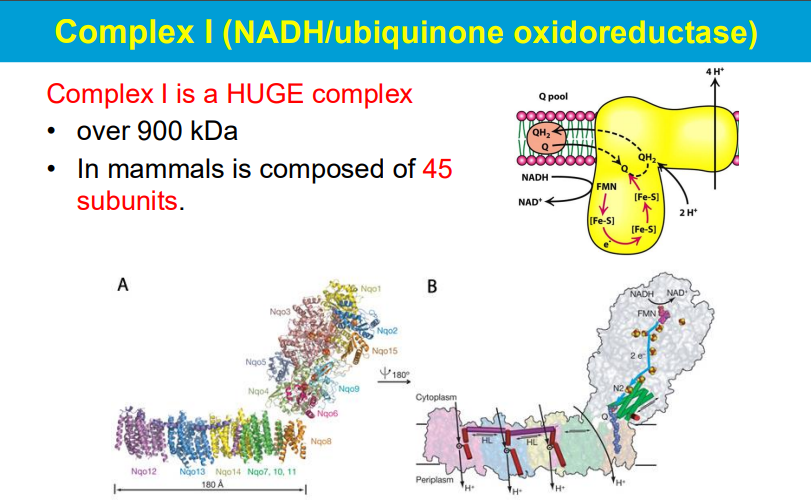
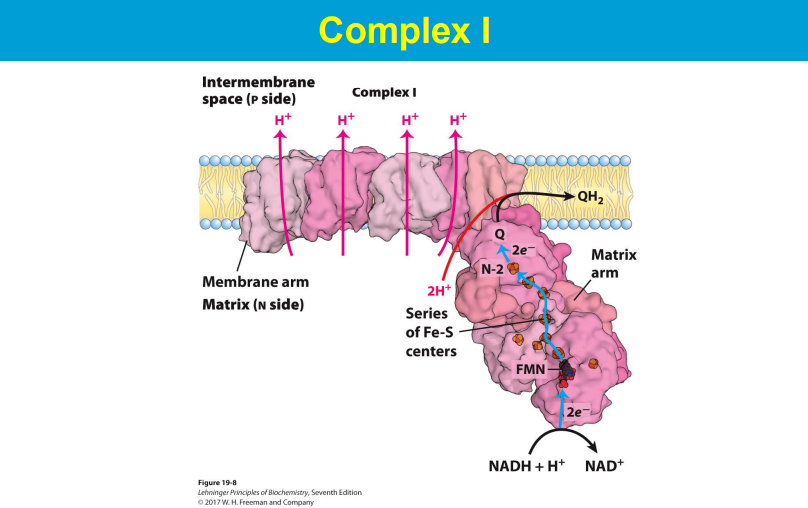
FMN accepts 2 e- then donates one at a time to Q
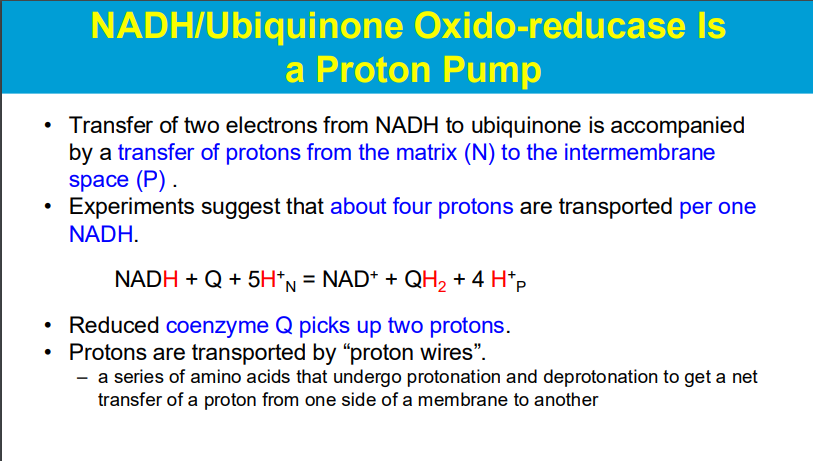
by donating 2 e-, NADH drives the 4 H+ to be pumped out into the IMS
2 e- are donated to Q/ubiquinine, and then Q picks up 2 H+ via proton wires
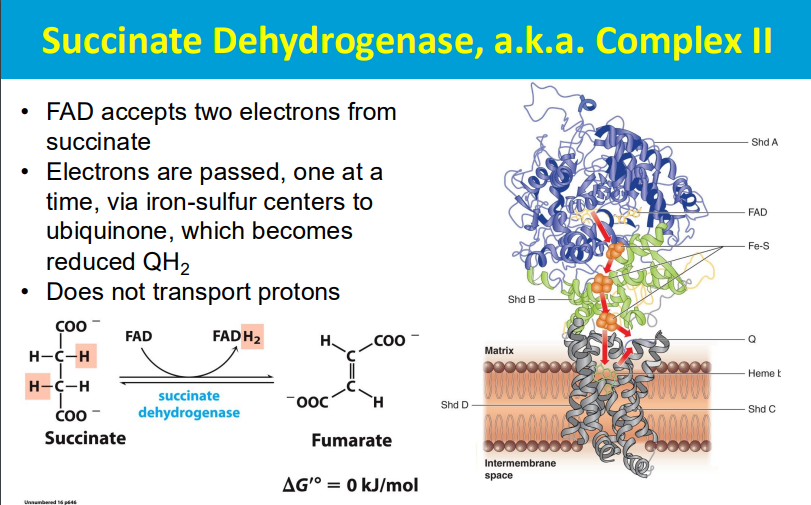
DOES NOT TRANSPORT PROTONS
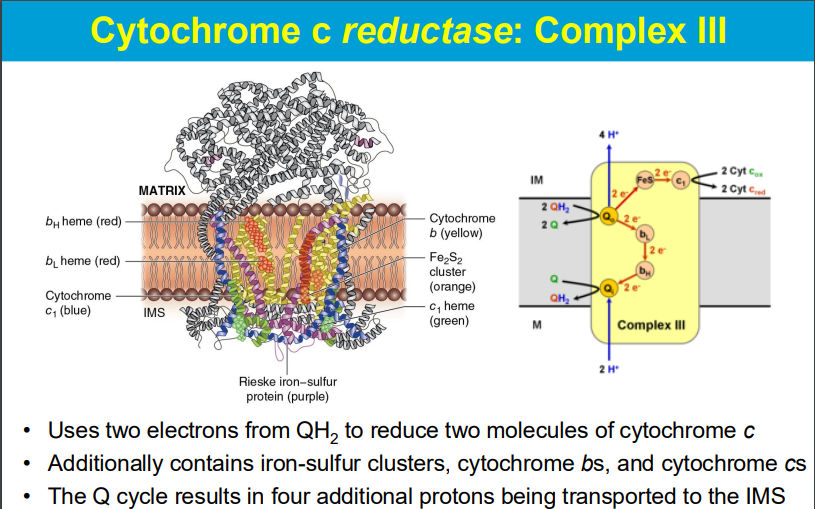
QH2 (mobile electron transporter) from complex I and II
**DOES TRANSPORT PROTONS
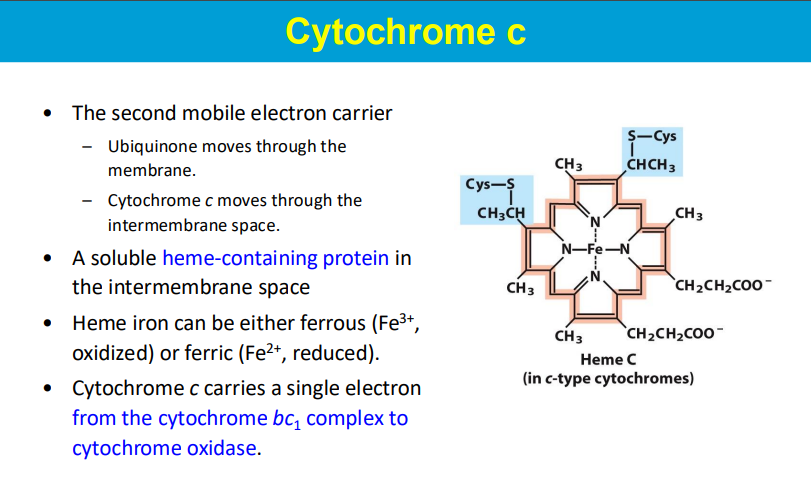
another mobile electron carrier (carries 1 electron)
carries e- to complex 4

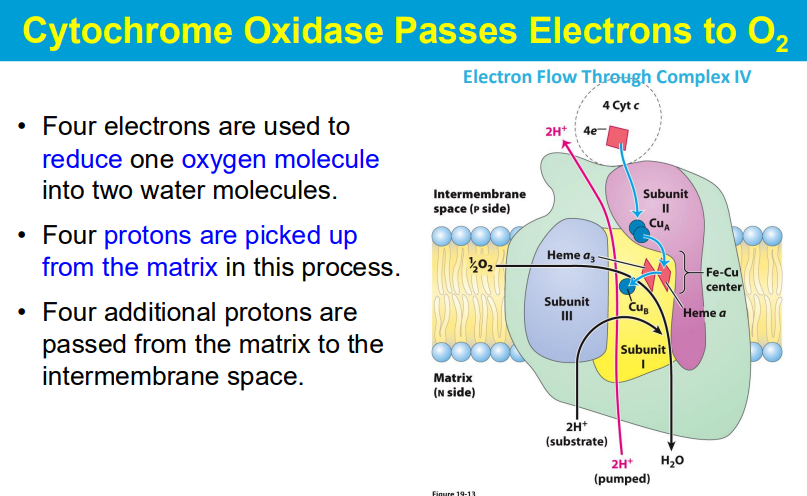
needs 4 e- to reduce O2 into 2H20
2 e- to reduce 1/2O2 into 1 H20
PROTON TRANSFER
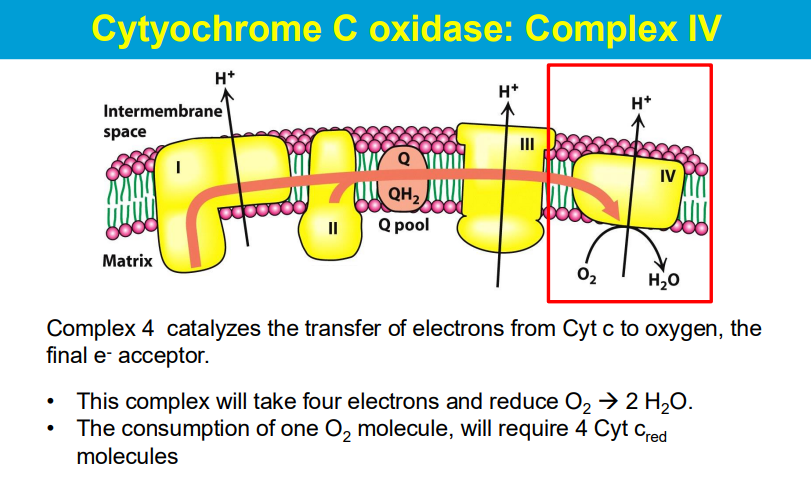
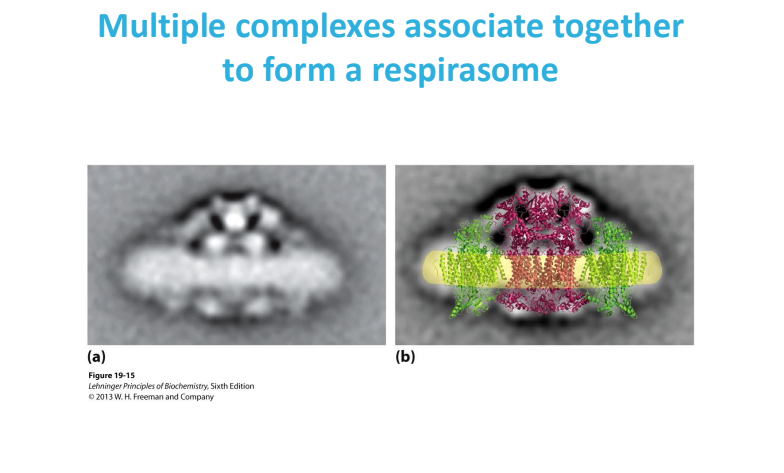
A respirasome is a large molecular machine in the inner mitochondrial membrane that transfers electrons from donors to oxygen and is a key part of the oxidative phosphorylation system
a respirasome is a functional grouping of ETC complexes, while the ETC is the individual components that make up the electron transfer pathway itself
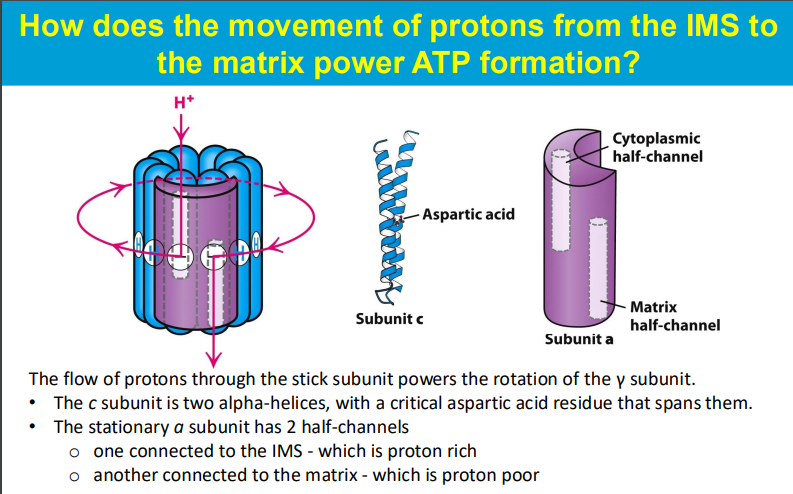
The movement of protons from the intermembrane space (IMS) to the mitochondrial matrix powers ATP formation through the following mechanism:
As protons flow down their concentration gradient from the IMS (where they are in higher concentration) into the mitochondrial matrix (where they are in lower concentration), they pass through the ATP synthase enzyme, specifically through the c subunit. The c subunit consists of multiple alpha-helices, with a critical aspartic acid residue that plays a key role in this process.
This proton flow causes the c subunit to rotate, which in turn powers the rotation of the γ subunit of ATP synthase. The rotation of the γ subunit induces conformational changes in the β subunits of ATP synthase, leading to the binding of ADP and inorganic phosphate (Pi) and facilitating their conversion into ATP.
The stationary a subunit of ATP synthase contains two half-channels: one that opens to the IMS (proton-rich) and another that opens to the matrix (proton-poor). Protons enter the a subunit from the IMS, bind to the c subunit, and as they pass through, they cause the rotation necessary for ATP synthesis. This entire process is an excellent example of chemiosmosis, where the energy stored in the proton gradient is converted into chemical energy in the form of ATP.
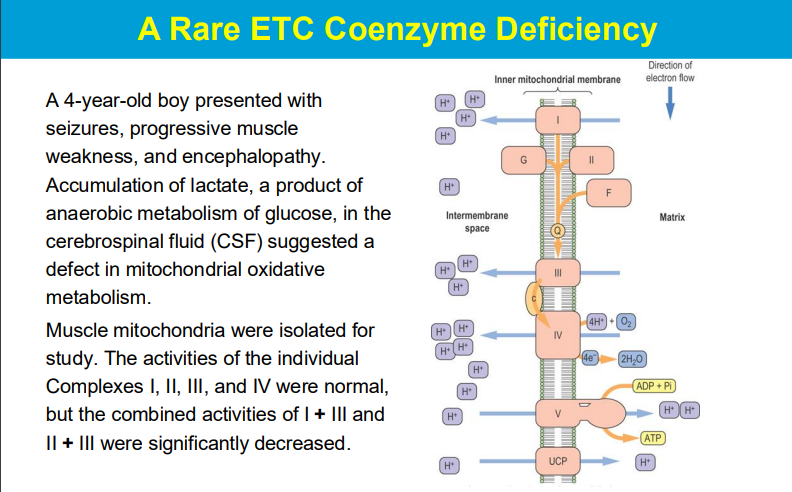
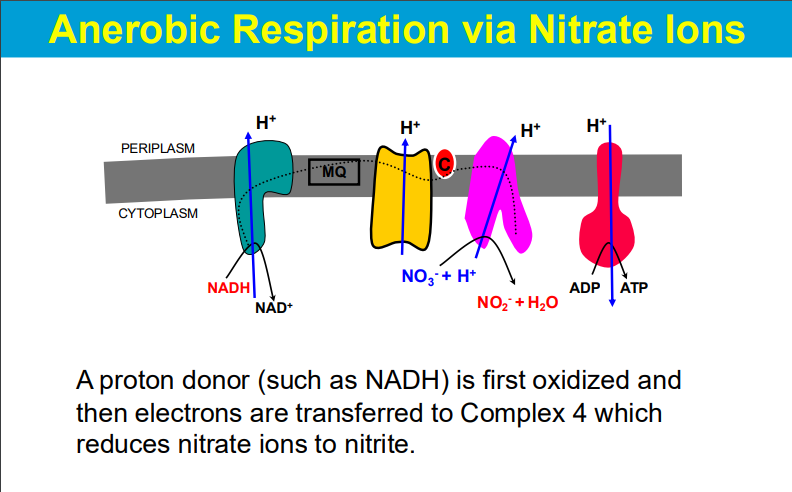
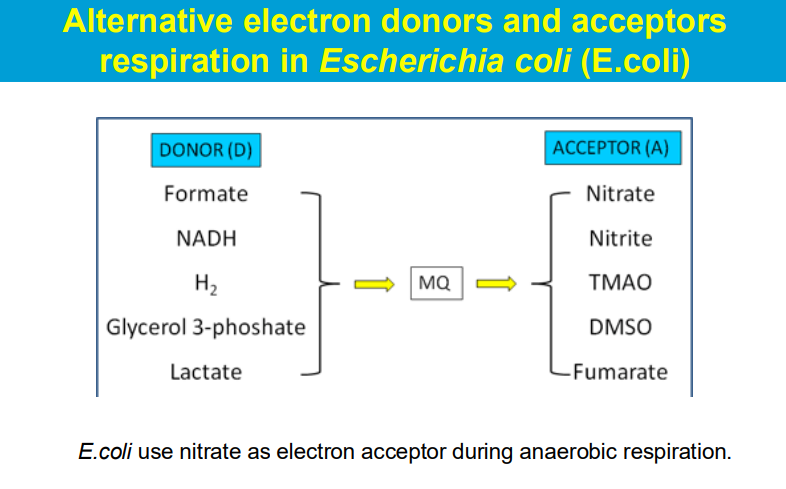

O2 is not the only final electron acceptor
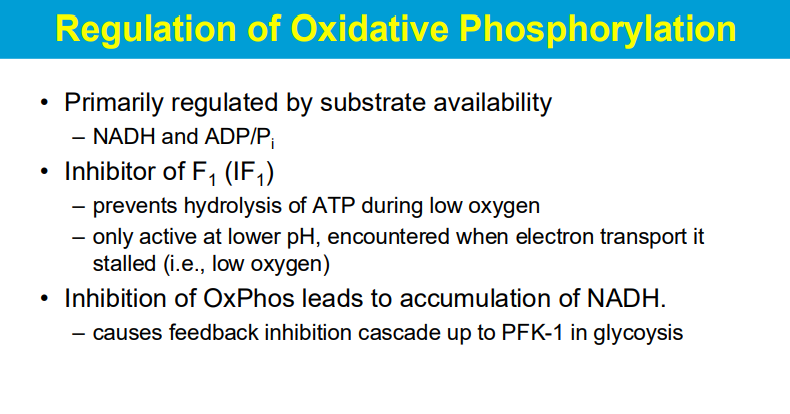
"Inhibitor of F1 (IF1)" refers to a protein that specifically prevents the breakdown of ATP (hydrolysis) when oxygen levels are low, and it becomes active only under acidic conditions (lower pH) which typically occur when the electron transport chain is stalled due to lack of oxygen; essentially acting as a protective mechanism to conserve ATP during low oxygen situations.
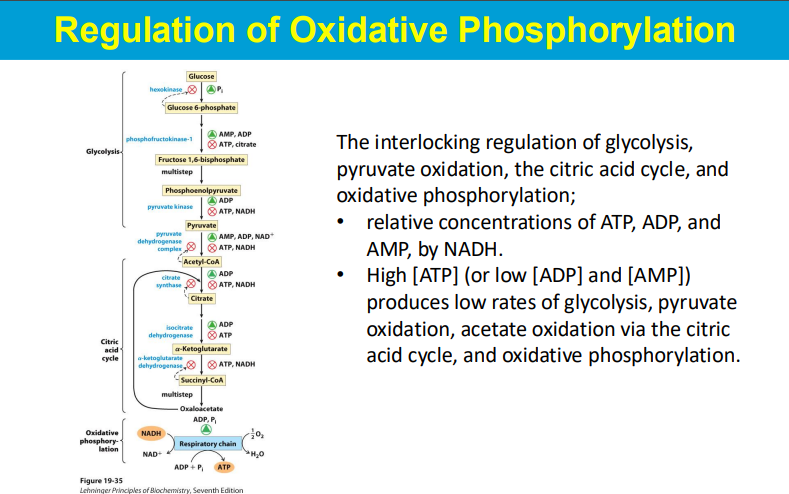
When oxidative phosphorylation (OxPhos) is inhibited, the resulting build-up of NADH within the cell triggers a feedback inhibition cascade that ultimately impacts the activity of phosphofructokinase-1 (PFK-1), a key regulatory enzyme in glycolysis, effectively slowing down the glycolytic pathway
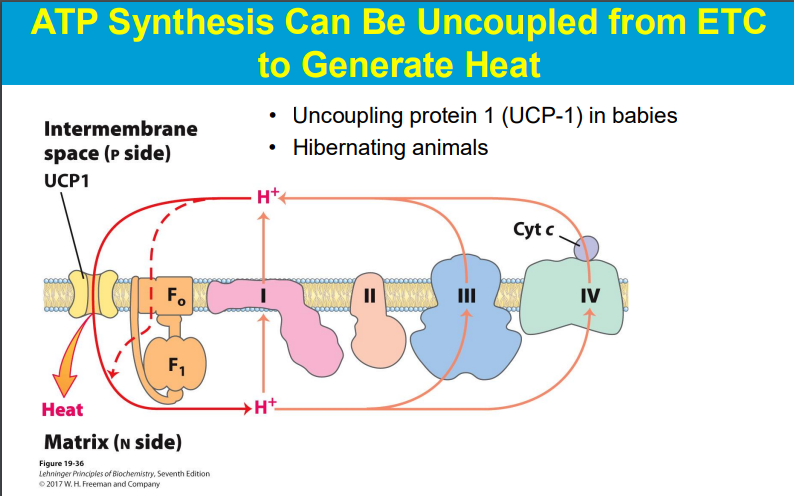
a process called "oxidative phosphorylation uncoupling" where the energy normally used to produce ATP is instead released as heat due to the leakage of protons across the mitochondrial membrane, bypassing ATP synthase
Oxidative phosphorylation uncoupling happens in babies primarily because of the presence of "brown adipose tissue" (BAT), which utilizes a protein called thermogenin (also known as uncoupling protein 1) to generate heat through a process called non-shivering thermogenesis, crucial for maintaining body temperature in newborns who have a large surface area relative to their body volume and are prone to heat loss; this uncoupling essentially allows energy to be dissipated as heat instead of being used to produce ATP.
babies have more UCP-1 → produce heat from proton motive force instead of ATP
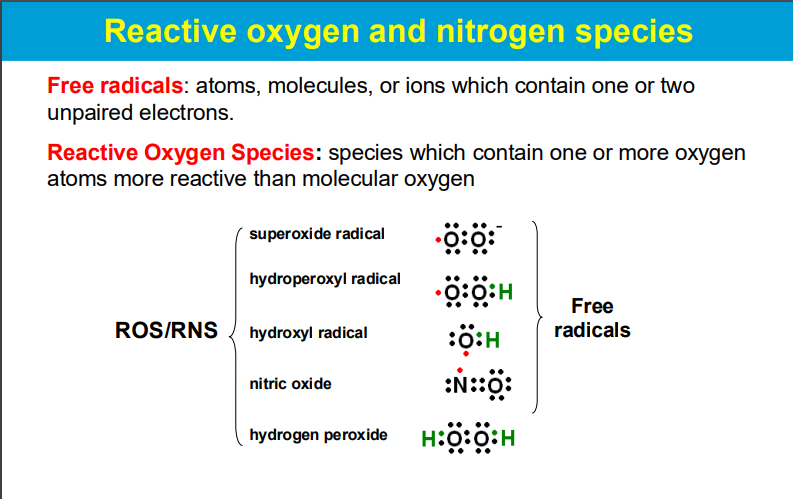
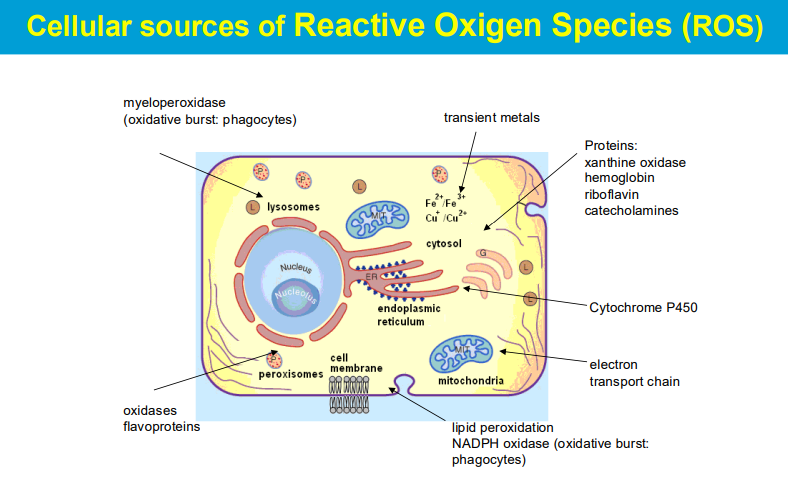
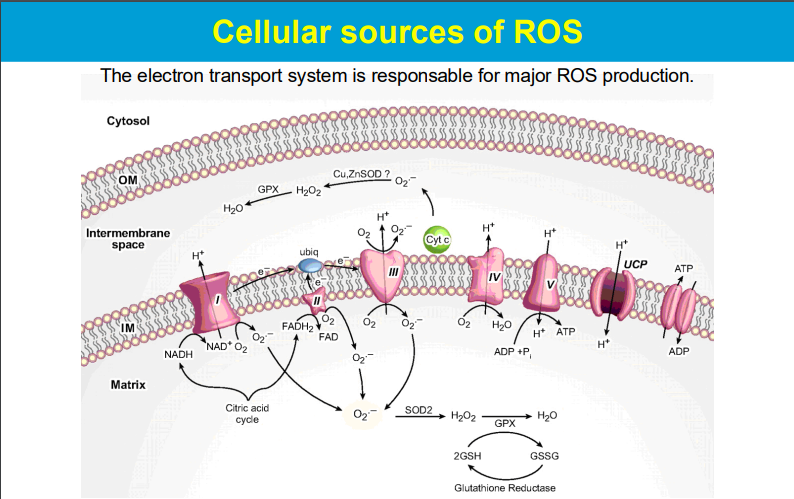
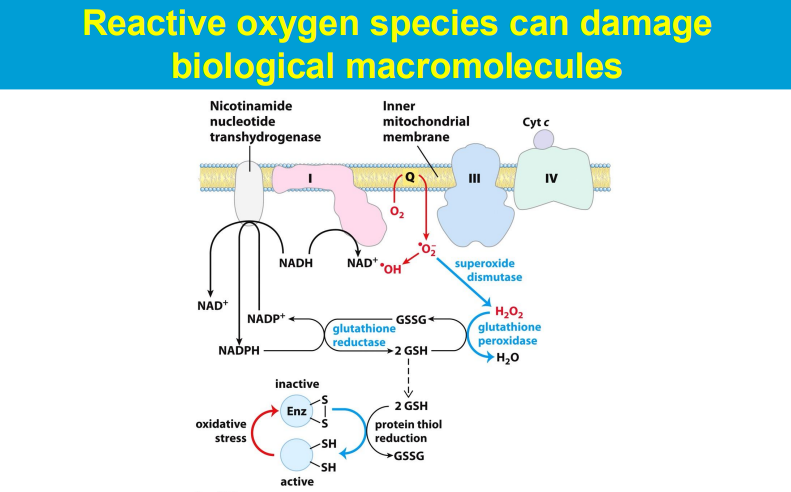
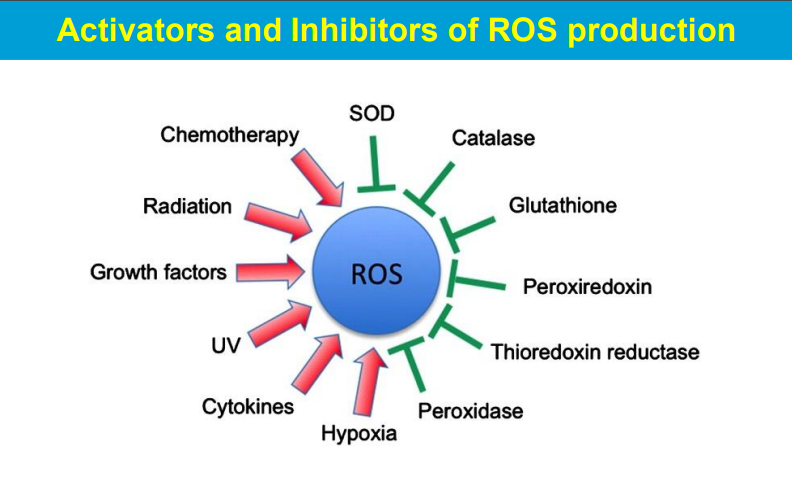
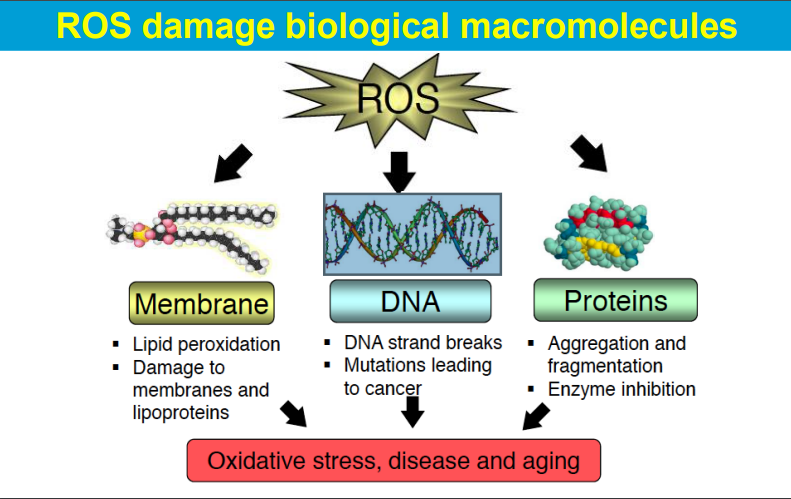
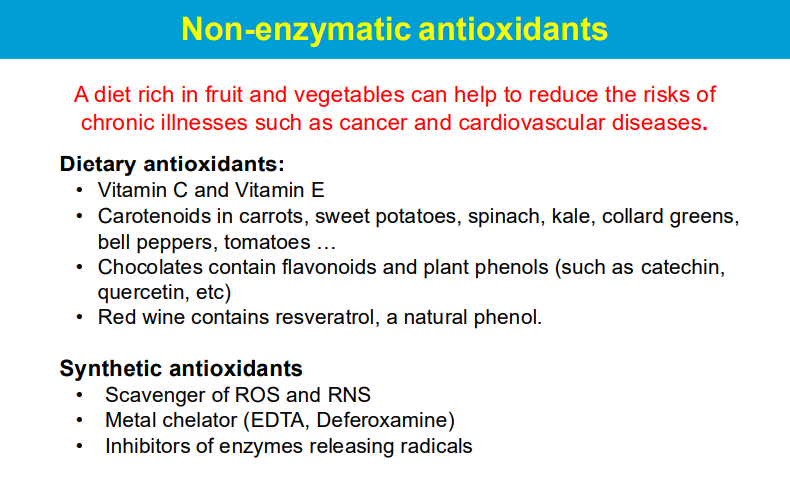
causing significant harm to cells and tissues when produced in excess, leading to a condition known as oxidative stress; this damage can contribute to various diseases and aging processes.
ROS are highly reactive molecules with unpaired electrons that can readily react with other molecules, causing chemical modifications and disrupting their normal function.
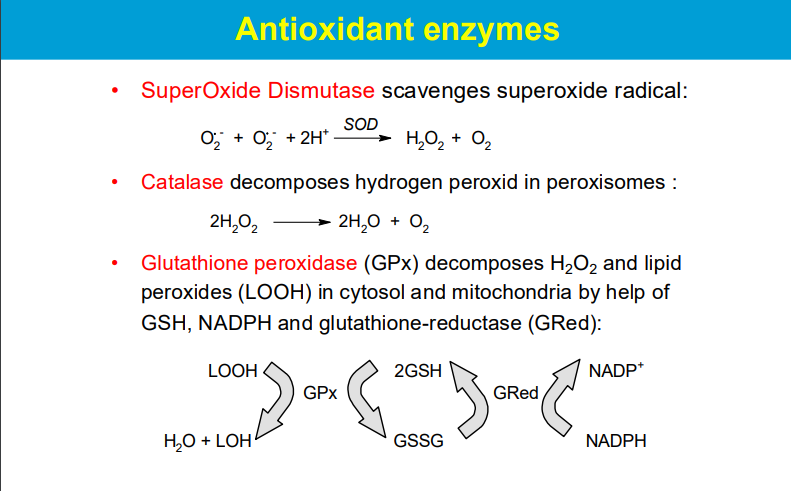
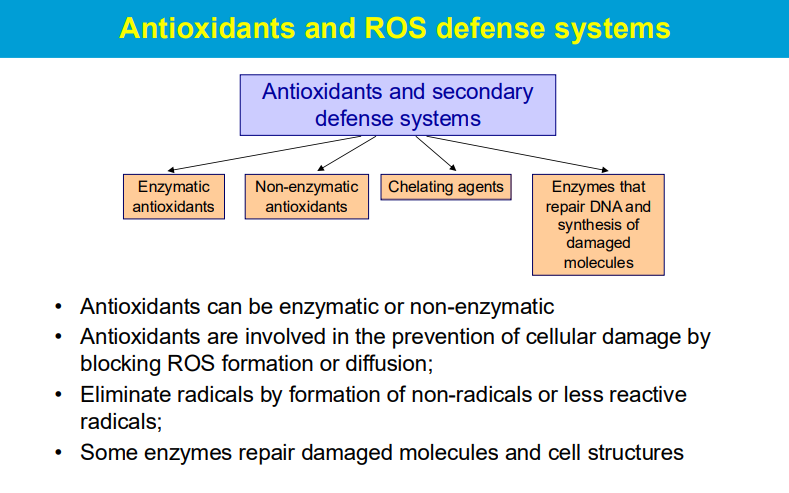
Superoxide Dismutase (SOD):
This enzyme converts the highly reactive superoxide radical (O2-) into hydrogen peroxide (H2O2) and molecular oxygen (O2), effectively neutralizing the superoxide radical.
Catalase:
Located mainly in peroxisomes, catalase rapidly decomposes hydrogen peroxide into water and oxygen.
Glutathione Peroxidase (GPx):
This enzyme uses glutathione (GSH) as a reducing agent to convert hydrogen peroxide and lipid peroxides into harmless products, with the oxidized glutathione (GSSG) being recycled back to GSH by glutathione reductase (GRed) using NADPH as an electron donor.
**While SOD is the first line of defense against superoxide radicals, the subsequent breakdown of the produced hydrogen peroxide relies on enzymes like catalase and glutathione peroxidase.
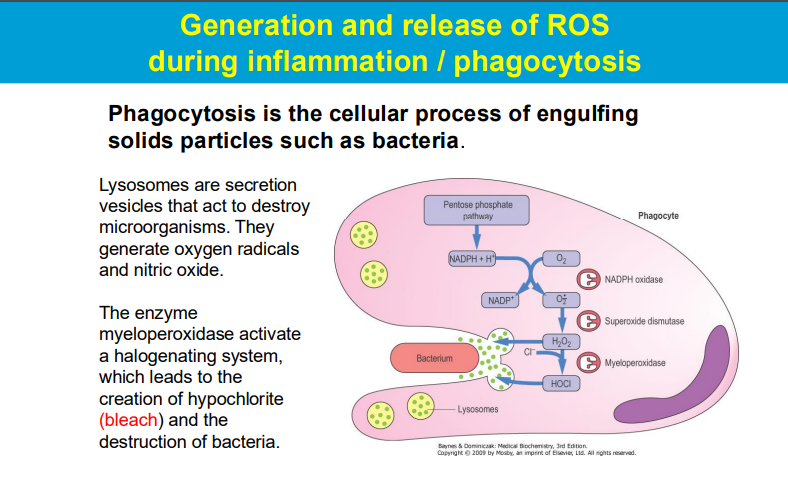
Lysosomes in Phagocytosis: Lysosomes are specialized organelles that contain digestive enzymes and are involved in the breakdown of microorganisms that are engulfed by immune cells (such as macrophages and neutrophils) during phagocytosis. When a pathogen is phagocytosed, it is enclosed in a phagosome that fuses with lysosomes, forming a phagolysosome.
Generation of ROS: During the immune response, especially during inflammation, activated phagocytes generate ROS. These reactive molecules, including superoxide anions, hydrogen peroxide, and hydroxyl radicals, play a crucial role in killing bacteria and other pathogens.
Myeloperoxidase and Halogenation: The enzyme myeloperoxidase (MPO), found in neutrophils, catalyzes the conversion of hydrogen peroxide into hypochlorite (bleach) in the presence of chloride ions. This hypochlorite is a potent antimicrobial agent that helps destroy bacteria and fungi.
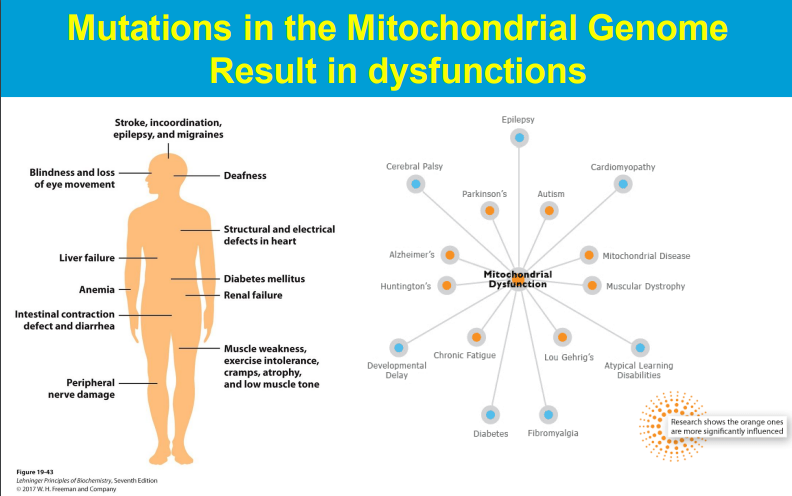

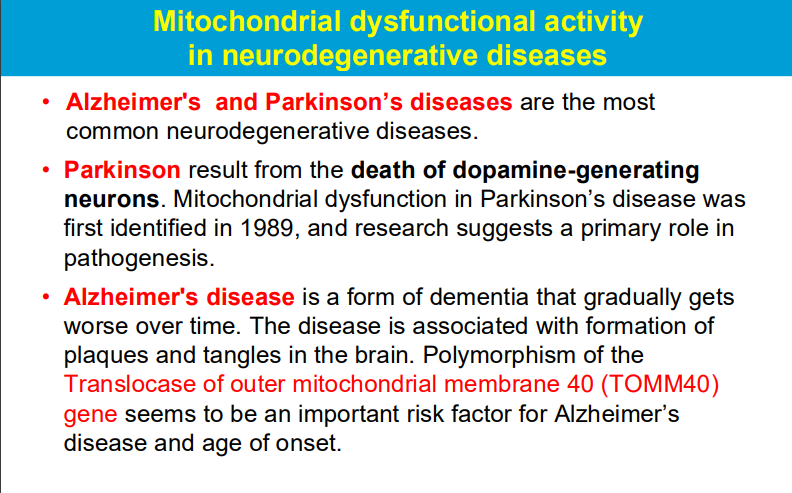
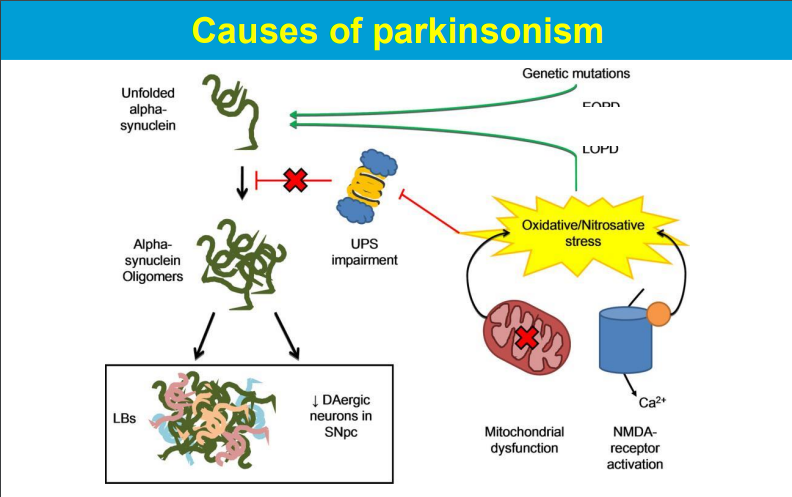
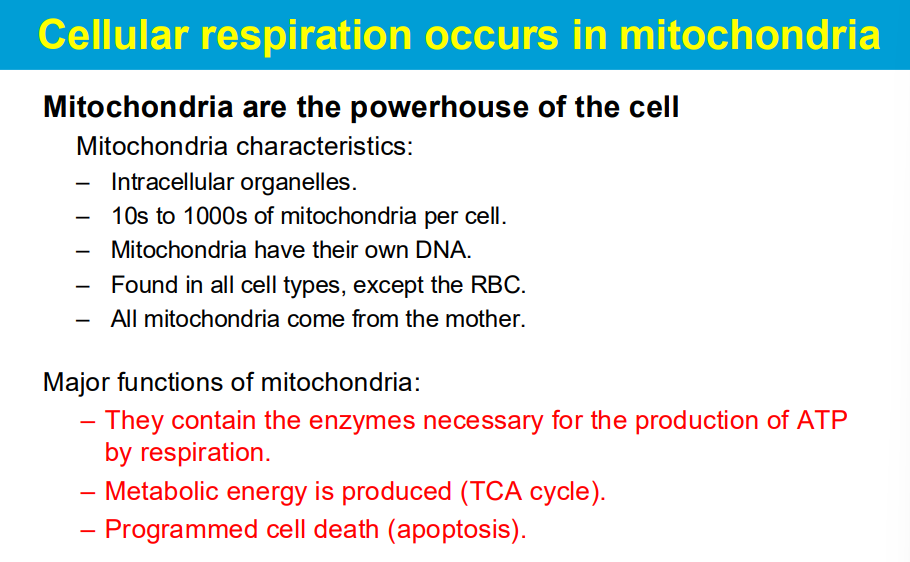
Free radicals produced during mitochondrial respiration can damage mitochondrial components including DNA, leading to impaired function and contributing to disease development.