EXAM 4 LO
Learning Objectives
DNA Replication
Illustrate semi-conservative replication with drawings.
Draw a Map to avoid “mind fields”
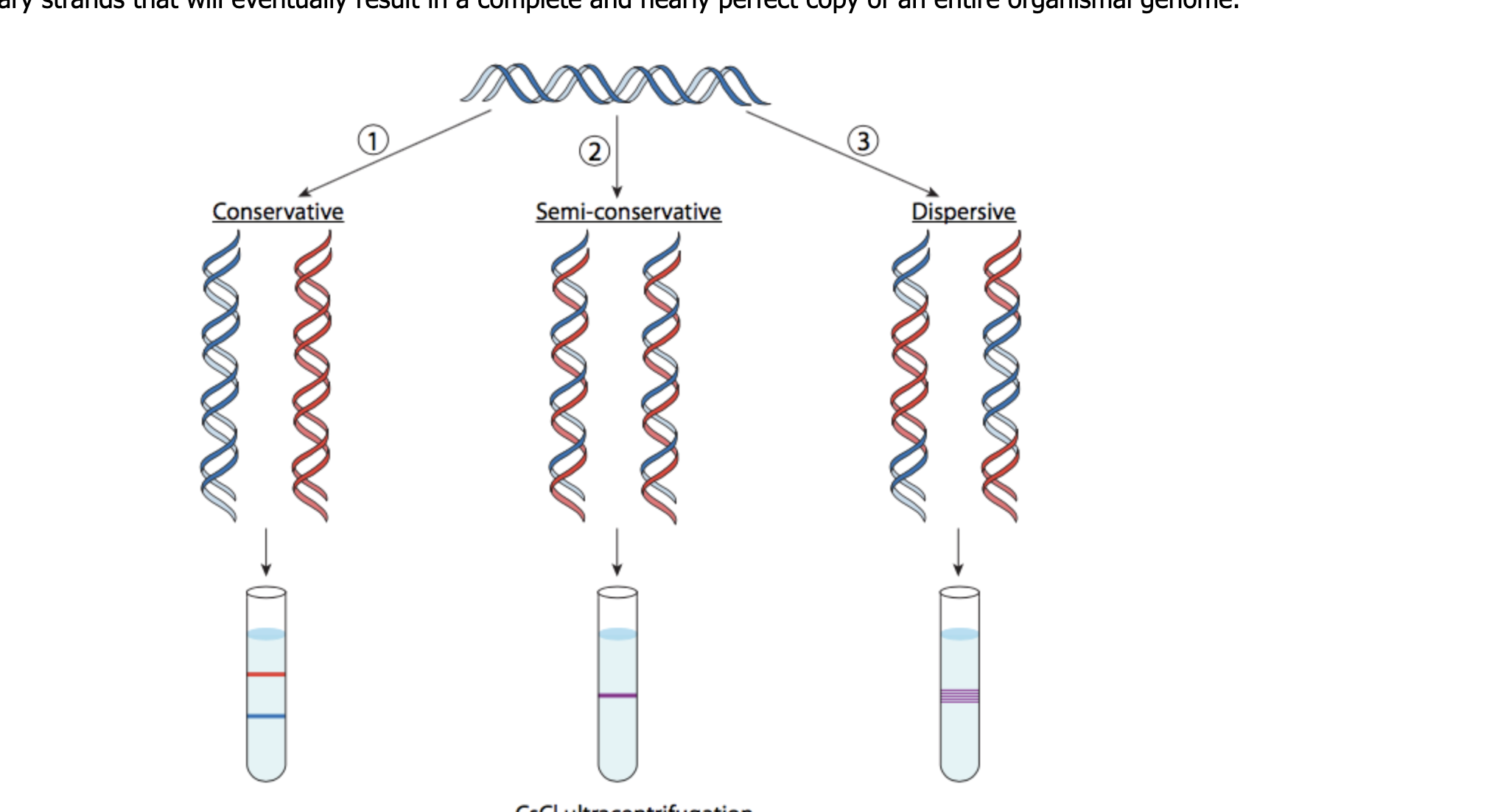
Explain Meselson and Stahl’s experiment results supporting semi-conservative replication, exploring three hypotheses:
Hypothesis 1: Semiconservative replication
States each strand of the original DNA molecule serves as a template for the production of a new complementary strand. Therefore, after replication, each new DNA molecule consists of one old strand and one new strand, preserving the parental DNA sequence across generations.
Hypothesis 2: Conservative replication
States the original DNA double helix remains intact, and a completely new DNA helix is synthesized. This would imply that after replication, one molecule would retain both strands of the original DNA and the other molecule would possess two newly synthesized strands, which ultimately would not match the original helix.
Hypothesis 3: Dispersive replication
States that the original DNA strands break down into pieces, and during replication, these pieces are reassembled to form two new double helices. This would result in each strand of the new DNA containing segments of both old and new DNA, leading to a mosaic of parental and daughter DNA incorporated into each strand.
Clarify the role of RNA polymerase in DNA replication.
The function of RNA polymerase in DNA replication is to synthesize RNA from a DNA template.
It unwinds DNA and adding complementary RNA nucleotides, thereby generating messenger RNA (mRNA) that carries the genetic information required for protein synthesis.
Draw and label origin of replication, replication forks, continuous vs. discontinuous DNA synthesis.
Explain telomerase activity and its role at chromosome ends, identifying it as an RNA-dependent DNA polymerase.
Telomerase is an enzyme with an essential role at the ends of chromosomes, known as telomeres. It is classified as an RNA-dependent DNA polymerase because it uses an RNA template to synthesize DNA.
Telomerase adds repetitive nucleotide sequences to the telomeres, protecting chromosome ends from degradation and allowing for complete DNA replication helping to maintain chromosomal integrity and stability.
Compare cellular DNA replication with PCR processes.
Purpose:
Cellular DNA replication serves to duplicate the entire genome of a cell prior to cell division, ensuring that each daughter cell receives a complete set of genetic information.
PCR is a laboratory technique used to amplify specific segments of DNA, making millions of copies of a targeted DNA sequence for analysis or experimentation.
Location:
Cellular DNA replication occurs within the cell nucleus in eukaryotes and within the cytoplasm in prokaryotes.
PCR takes place in a controlled laboratory environment, often using thermal cyclers.
Enzymes:
In cellular DNA replication, enzymes such as DNA polymerases, helicases, and ligases are involved in the replication process.
In PCR, a specialized DNA polymerase (commonly Taq polymerase) is used, which is heat-stable and allows for the denaturation and re-annealing of DNA strands at high temperatures.
Nucleotides:
During cellular DNA replication, deoxynucleotides (dNTPs) are added to the growing DNA strands.
In PCR, the same deoxynucleotides (dNTPs) are used, but are added during cycles of denaturation, annealing, and extension, allowing for amplification.
DNA Transcription
Contrast DNA replication and transcription in terms of purpose, location, enzymes, and nucleotides.
Purpose:
DNA Replication: To duplicate the entire genome of a cell before cell division, ensuring each daughter cell receives a full set of genetic information.
Transcription: To synthesize RNA from a DNA template, producing messenger RNA (mRNA) that carries genetic information for protein synthesis.
Location:
DNA Replication: Occurs in the cell nucleus in eukaryotes and the cytoplasm in prokaryotes.
Transcription: Takes place in the cell nucleus for eukaryotes, while in prokaryotes, it occurs in the cytoplasm as well.
Enzymes:
DNA Replication: Involves various enzymes like DNA polymerases, helicases, and ligases.
Transcription: Primarily involves RNA polymerase, which synthesizes RNA from the DNA template.
Nucleotides:
DNA Replication: Utilizes deoxynucleotides (dNTPs) during the replication process.
Transcription: Uses ribonucleotides (rNTPs) to form RNA, which differs from the nucleotides used in DNA replication.
Identify the DNA sequences for transcription initiation and termination.
DNA has special parts that tell it when to start and stop. These parts are like the beginning and end of a story. The start part is called the 'initiation sequence', and the end part is called the 'termination sequence'.
When the cell wants to make a message from the DNA, it looks for these special parts. The cell knows to start reading the DNA at the start part, and when it sees the end part, it knows to stop. It's like starting to read a book and then knowing when to close it when you're done!
Explain the DNA template, non-template, and coding strands.
Template Strand: This is the strand of DNA that serves as the actual template for RNA synthesis during transcription. RNA polymerase reads this strand in the 3' to 5' direction, synthesizing the new RNA strand in the 5' to 3' direction. The sequence of the RNA will be complementary to this strand.
Non-template Strand (also known as the sense strand): This strand is not directly used for transcription but has the same sequence as the RNA produced (except for the substitution of uracil for thymine in RNA). It is called the non-template strand because it does not serve as a template for RNA synthesis.
Coding Strand: Another name for the non-template strand, the coding strand has the same sequence as the RNA transcript produced from the template strand, making it useful in predicting the RNA sequence without the need for transcription.
Discuss pre-mRNA processing in eukaryotes, including components of introns and exons and the relevance of alternative splicing.
Components of Introns and Exons:
Introns: These are non-coding sequences found within a gene that are transcribed into pre-mRNA but are removed during processing. Introns can vary in size and often contain regulatory elements that may influence gene expression.
Exons: These are coding sequences that remain part of the mature mRNA after processing. They carry the actual information which is translated into proteins. Exons can occur in various combinations, contributing to protein diversity.
Relevance of Alternative Splicing:
Alternative splicing is a crucial process that allows a single pre-mRNA transcript to be spliced in different ways, resulting in multiple mRNA isoforms.
This enhances the versatility of gene expression, as different combinations of exons can lead to the production of distinct proteins from a single gene.
This mechanism is vital for increasing the functional complexity of the proteome without necessitating a corresponding increase in the number of genes. Alternative splicing is also associated with various biological processes and can impact development, tissue-specific functions, and cellular responses.
DNA Translation
• Explain the role of tRNA molecules and aminoacyl tRNA synthetases
tRNA molecules are crucial for protein synthesis as they transport amino acids to the ribosome, matching them with the appropriate codon on the mRNA. Aminoacyl tRNA synthetases are enzymes responsible for catalyzing the attachment of specific amino acids to their corresponding tRNA molecules, ensuring that the correct amino acids are incorporated into the growing polypeptide chain based on the mRNA sequence.
• Describe where codons and anticodons are found and their role
Codons are found on mRNA and consist of three nucleotide bases that specify amino acids during protein synthesis. Anticodons are located on tRNA and contain three complementary bases to the codon, allowing tRNA to correctly match the amino acid to the corresponding codon on the mRNA, facilitating the translation process.
• Remember ATG/AUG as the start and methionine codon, and that stop codons are recognized by a protein release factor (not a tRNA)
ATG/AUG is the start codon that codes for methionine. Stop codons are recognized by a protein release factor instead of a tRNA.
• Explain the concept of reading frame and properly use a genetic code chart/table
The reading frame is a way of dividing the sequence of nucleotides in DNA or RNA into groups of three, known as codons, which correspond to amino acids.
Proper use of a genetic code chart/table involves aligning the mRNA sequence with the appropriate codons to determine the resultant polypeptide chain during translation.
• Explain how the genetic code is redundant, yet not ambiguous
The genetic code is described as redundant because multiple codons can specify the same amino acid;
For example, both GAA and GAG code for glutamic acid. However, it is not ambiguous because each codon corresponds to only one specific amino acid, ensuring that the genetic instructions remain clear and precise in protein synthesis.
• Explain and distinguish the two mechanisms resulting in genetic code redundancy
The genetic code is redundant: multiple codons can code for the same amino acid (e.g., GAA and GAG both code for glutamic acid), but it is not ambiguous, as each codon corresponds to only one specific amino acid.
• Describe and distinguish silent (synonymous), missense, nonsense, and frameshift mutations
Types of Mutations:
Silent (Synonymous) Mutations: Changes in DNA sequence that do not alter the amino acid sequence of a protein due to redundancy in the genetic code.
Missense Mutations: Changes that result in the substitution of one amino acid for another in the protein, potentially affecting its function.
Nonsense Mutations: Mutations that create a premature stop codon, leading to a truncated and usually nonfunctional protein.
Frameshift Mutations: Insertions or deletions of nucleotides that shift the reading frame, altering the entire amino acid sequence downstream, often resulting in a completely different and nonfunctional protein.
Nucleotides
Describe the general structure of nucleosides and nucleotides, distinguishing those of DNA from those of RNA, remembering which are purines and which are pyrimidines
Nucleosides consist of a nitrogenous base (either a purine or a pyrimidine) attached to a sugar (ribose in RNA, deoxyribose in DNA). Nucleotides, the building blocks of nucleic acids, include a nucleoside and one or more phosphate groups. The key purines are adenine (A) and guanine (G), while the pyrimidines are cytosine (C), thymine (T, in DNA), and uracil (U, in RNA).
Remember what NTP and dNTP stand for, and which bases are included in each
NTP (nucleoside triphosphate) refers to the building blocks of RNA, which include adenine (A), cytosine (C), guanine (G), and uracil (U). dNTP (deoxynucleoside triphosphate) refers to the building blocks of DNA, composed of adenine (A), cytosine (C), guanine (G), and thymine (T).
Remember the significance of the 1’, 2’, 3’ and 5’ positions of ribose and deoxyribose
The 1’ position connects to the nitrogenous base, crucial for base pairing.
The 2’ position distinguishes RNA from DNA due to the presence of a hydroxyl group in RNA, impacting its stability.
The 3’ position allows the linking of nucleotides, maintaining directionality in nucleic acid synthesis.
The 4’ position, while not as commonly discussed, contributes to the sugar's structural integrity, ensuring proper conformation of the nucleotide.
Lastly, the 5’ position attaches the phosphate group, vital for energy transfer and the polymerization of nucleic acids.
Recognize the 3’ → 5’ or 5’ → 3’ directionality of a nucleotide and nucleic acid
To recognize the directionality of nucleotides and nucleic acids, look for the phosphate group attached to the 5' carbon of the sugar. The direction proceeds from this 5' end to the 3' end, which is where the hydroxyl group on the 3' carbon is located. Therefore, a nucleotide strand is synthesized in a 5' to 3' direction, with the 5' end indicating where the chain begins and the 3' end where it ends.
Explain why nucleic acid polymerization (synthesis) is always 5’ to 3’
Nucleic acid polymerization occurs in the 5’ to 3’ direction because the phosphate group of the incoming nucleotide attaches to the hydroxyl group on the 3’ carbon of the previous nucleotide. This creates a growing chain where synthesis can only proceed by adding nucleotides to the 3’ end.
Summarize the major discoveries of Chargaff, Franklin, and Watson & Crick
Erwin Chargaff: Discovered that DNA composition varies among species and established Chargaff's rules, noting that the amount of adenine (A) equals thymine (T) and the amount of guanine (G) equals cytosine (C).
Rosalind Franklin: Utilized X-ray diffraction to capture images of DNA, providing evidence of its helical structure and dimensions, critical for understanding DNA's form.
James Watson & Francis Crick: Developed the double-helix model of DNA structure in 1953, integrating Chargaff’s and Franklin’s findings to explain how genetic information is stored and replicated.
Protein Targeting
Precisely describe where in a cell amino acids are linked together in protein synthesis
Amino acids are linked together in the ribosome, a complex molecular machine found in all living cells. During protein synthesis, ribosomes can be found either freely floating in the cytosol or attached to the endoplasmic reticulum (ER), specifically the rough ER, giving it a studied 'rough' appearance. The process begins with messenger RNA (mRNA), which is transcribed from DNA and carries the genetic instructions necessary for constructing proteins.
Once the mRNA reaches the ribosome, it serves as a template guiding the sequence of amino acids that will be assembled into a polypeptide chain. Transfer RNA (tRNA) molecules then deliver specific amino acids to the ribosome, matching their anticodons to the corresponding codons on the mRNA strand. The ribosome facilitates the formation of peptide bonds between the amino acids, linking them together in the correct order as dictated by the mRNA sequence, ultimately resulting in a newly synthesized protein that will undergo further processing and folding before assuming its functional form.
Define a “secretory” protein as a cell biologist would
Secretory proteins are those synthesized in cells to be released outside or function at the plasma membrane. They differ from non-secretory proteins based on their trafficking; secretory proteins are directed towards the endoplasmic reticulum (ER) for further modifications and are ultimately secreted, while non-secretory proteins may remain in the cytosol or transit to other organelles.
Co-translational translocation refers to the process where emerging polypeptides enter the ER during synthesis. Mature proteins often lack the initiating methionine due to cleavage during processing. Modifications in the ER lumen include glycosylation and folding, influencing whether a protein is secreted or retained as an integral membrane protein.
Explain what distinguishes “secretory” from “non-secretory” proteins and how that difference regulates protein trafficking in our cells
Secretory proteins are synthesized to be released outside the cell or function at the plasma membrane, requiring trafficking to and processing in the endoplasmic reticulum (ER). In contrast, non-secretory proteins remain in the cytosol or other organelles, not directed toward secretion.
During synthesis, secretory proteins undergo co-translational translocation into the ER, while common post-translational modifications can occur there, affecting their final destination. The distinction between secretory and non-secretory proteins regulates their trafficking and determines whether they are secreted or retained in cellular compartments.
Explain or draw what is meant by co-translational translocation of polypeptides
in notebook
Explain why many mature proteins lack methionine at their amino terminus, even though methionine is encoded by start codons
Many mature proteins lack methionine at their amino terminus despite methionine being encoded by start codons because the initiating methionine can be cleaved off during post-translational modifications within the endoplasmic reticulum (ER) or during further processing of the protein.
Post-translational modification refers to the processes that occur after a protein has been synthesized, which can lead to the addition or removal of various chemical groups or moieties. These modifications can include the cleavage of the initiating methionine, glycosylation, phosphorylation, and other alterations that influence the protein's activity, stability, localization, and overall function.
Describe common modifications to proteins within the ER lumen
Common modifications to proteins within the ER lumen include glycosylation, which adds carbohydrate groups, and folding, which ensures proper protein conformation. Additional modifications can involve disulfide bond formation, which stabilizes the protein structure, and proteolytic cleavage, which activates the protein by removing segments. These modifications are crucial for protein functionality and trafficking.
Methionine, coded by the start codon (AUG), serves as the initial amino acid in the synthesis of proteins. It plays a crucial role in initiating the translation process, signaling the start of polypeptide formation in the ribosome. Oftentimes cleaved off during post-translational modification
Explain what determines whether a secretory protein will be literally secreted into the extracellular milieu or be retained as an integral protein at the plasma membrane
The determination of whether a secretory protein is secreted into the extracellular space or retained as an integral membrane protein involves several key factors. First, specific signal sequences within the protein's amino acid sequence act as molecular tags that guide the protein to its appropriate destination. These signal sequences can direct the protein to the endoplasmic reticulum (ER) for further processing and packaging.
Describe where the intracellular and extracellular domains of a transmembrane protein exist before that protein reaches the plasma membrane
Before reaching the plasma membrane, the intracellular domain of a transmembrane protein exists within the cytoplasm of the cell, while the extracellular domain is exposed to the outside environment. This arrangement allows for the protein to interact with both the internal cellular machinery and external signals.
Explain what determines whether a non-secretory protein will remain in the cytosol, traffic to the nucleus, or be imported into another intracellular compartment
The fate of a non-secretory protein—whether it remains in the cytosol, traffics to the nucleus, or enters another intracellular compartment—is determined by specific signal sequences within its amino acid chain. These sequences act as molecular tags that guide the protein to its designated location within the cell.