AP Bio: Evolution (unit 7)
HISTORY OF LIFE ON EARTH
conditions of early earth: reducing/electron-adding atmosphere that contained CO2, CH4, NH3, H2O, N2, and H2
elements needed for life: C H O N
sources of fossils: sedimentary rocks, amber, mammals frozen in soil
the fossil record is incomplete because: 1) organisms didn’t all die in the right place and time to be fossilized correctly; 2) we’ve only discovered a small portion of all the fossilized organisms throughout history; 3) fossilization favors organisms that have certain characteristics; 4) some fossils are lost to natural and human causes
half-life: the time needed for a parent isotope (an unstable radioactive isotope that decays into stable daughter isotopes) to decay by 50%
radiometric dating: a technique that determines the age of a material by measuring the amount of radioactive isotope in it and comparing it to the amount of its decay products
continental drift: the phenomena where the landmasses that made up Pangaea drifted apart over time
miller-urey experiment: a closed-system experiment set up by Stanley Miller to emulate conditions on early Earth. miller ended up identifying a variety of organic molecules that are common in organisms (formaldehyde [CH2O], hydrogen cyanide [HCN], amino acids, and hydrocarbon chains). the result provided evidence that biomolecules CAN arise spontaneously from the conditions of the atmosphere of the early Earth.
haploid gametes: reproductive cells that contain a single set of chromosomes ( ½ the number of chromosomes found in normal body cells)
haploid: a cell that contains a single set of chromosomes
diploid: a cell containing 2 sets of chromosomes
protobionts/protocells: the first formed cells (aggregates of abiotically produced organic molecules surrounded by a membrane). seen as the precursors of cells
protists: simple eukaryotic organisms that are neither plants, animals, or fungi
the Cambrian Explosion: a period of rapid evolution, caused by UV light (due to the slow-forming ozone layer) and subsequent mutations.
pre-cambrian: aggregate multicellularity (clumps; no differentiation)
cambrian: true multicellularity with differentiation
emergence of modern invertebrates started here
endosymbiosis model:
a model that explains how organelles in eukaryotic cells evolved from “parasites” living in hosts to the organelles in the eukaryotes today. archaebacteria engulfed eubacteria that could perform aerobic respiration/photosynthesis
eubacteria that could do aerobic respiration → mitochondria
cyanobacteria → chloroplasts
evidence: mitochondria & chloroplasts reproduce independently, have their own DNA, and have a double membrane
endosymbionts (smaller cells) joined the cell by being eaten or as parasites. instead of being digested, the endosymbionts formed a symbiotic relationship with the larger cell. over time, the “organelle” and the host cell developed together (co-evolution) and the organelle was forever incorporated as an “organelle”
Steps to Modern Earth and its Life Forms:
biomolecular building blocks form
miller-urey experiment
amino acids, simple sugars, nucleotide precursors, fatty acids
without oxidation (from the presence of oxygen) complex biomolecules can survive and increase over time
membrane structures and cells emerge
protobionts
cell precursors; self-containment and a high concentration of biomolecules
RNA
acted like an enzyme; also self-replicating
inheritance and cell division
early protocells had no inheritance capability- just split into ½ to divide
RNA → store genetic info & replicate
haploid organisms
later became diploid…why is it good to be diploid?
→ 2 copies of the chromosome, so if anything goes wrong with one copy, you have backup! also, having 2 chromosomes means that the cell has greater genetic potential and can do more things
heterotroph → autotroph
earliest organisms: prokaryotes (single-celled anaerobic heterotrophs. absorbed nutrients)
photosynthesis → sugars created!
O2 production started, which increased over time in the atmosphere and killed most of the anaerobic organisms
aerobic respiration emerged; more energy per glucose molecule
eukaryotic & multi-celled organisms
eukaryotes (1 bil. years after prokaryotes)
compartmentalization
infolding membranes
organelles (complex metabolism)
3 domains: archaea, eukaryota, eubacteria
emergence of larger land masses
pangaea; continental drift → speciation and diversity
PHYLOGENY AND CLADISTICS
levels of classification: domain>kingdom>phylum>class>order>family>genus>species
species: a group of organisms that can breed and produce fertile offspring
taxon: a hierarchical classification level
phylogenetic tree: sorted by hierarchies; degrees of relatedness between 2 phyla can be seen by the # of categories separating them. the lower the category, the more related they are with a more recent ancestor
homologous similarities: phenotypic and genetic similarities due to shared ancestry. similar complex structures, bone structures, and DNA
analogous similarities: similarities between 2 organisms because of convergent evolution rather than shared ancestry. may look similar externally, but have different internal anatomy, physiology, etc.
parsimony/“occam’s razor”: the principle that the simplest explanation for a phenomenon is most likely to be true
3 domains: eukaryka, archaea, eubacteria
eukaryka: all organisms with cells containing true nuclei: includes single-celled organisms and multicellular plants, fungi, and animals (eukaryotes)
archaea: diverse groups of prokaryotic organisms that inhabit a wide variety of environments
eubacteria: most of the current known prokaryotes
INTRO TO EVOLUTION
Jean-Baptiste Lamarck: believed that traits acquired during your lifetime could be passed down to your offspring; theory of use and disuse; evolution happens because organisms have an inherent desire to be better
theory of use and disuse: parts of the body that are used extensively become bigger and stronger, while those that aren’t used as often deteriorate
the inheritance of acquired characteristics: organisms can pass on their modified characteristics to their offspring
descent with modification: evolution in which descent (shared ancestry, resulting in shared characteristics) and modification (the accumulation of differences) can be seen
theory of evolution by natural selection: organisms with traits that better adapt them to the environment are more likely to survive and reproduce, passing on their advantageous traits to their offspring. this leads to gradual changes in a population over generations, resulting in evolution.
homologous structures: variations on a structural theme that are present in a group of organisms
vestigial structures: remnants of features that served a function in the organism’s ancestors
comparative embryonic development: comparing early stages of development in different animal species to reveal anatomical homologies not seen in adult organisms. (many different species with common ancestors have similar-looking embryos!)
evolution trends: simple → complex; haploid → diploid; aquatic → terrestrial (long periods of increased diversity, and occasional periods of extinction)
evidence for evolution: biogeography, the fossil record (+index fossils), homologous structures, vestigial structures, comparative embryology
artificial selection: humans select for desired traits instead of natural ones
convergent evolution: independent evolution of similar features in different lineages. the organisms don’t have common ancestors but they end up having similar phenotypes
natural selection:
variation
competition for resources
fittest organisms
the “average” organism
mutations
EVOLUTION OF POPULATIONS
population: a group of organisms of the same species living in the same geographic area at the same time, producing fertile offspring (have a gene pool, and dom/rec allele frequencies)
mutations: a change in the usual DNA sequence of an organism, virus, or extrachromosomnal DNA
gene flow: the transfer of alleles between populations; immigration and emigration
genetic drift: chance events that alter allele frequencies (founder effect, bottleneck effect)
founder effect: when a few individuals become isolated from a larger population, they may establish a new gene pool and population that’s different from the source population
bottleneck effect: when a population passes through a “bottleneck” that reduces its size (sudden environment change, natural disaster, etc.)
directional selection: favors variants at one extreme of the distribution
disruptive selection: favors variants at both ends of the distribution
stabilizing selection: removes extreme variants from the population and preserves intermediate types
nonrandom/assortative mating: selecting a mate based on phenotype, giving that phenotype an advantage
sexual selection: in species where females prefer certain traits in their mates; a subversion of nonrandom mating (leads to sexual dimorphism)
sexual dimorphism: different phenotypes for each gender (see: peacocks, mallard ducks, lions, etc.)
divergent evolution: a process that occurs when a species or population splits into multiple groups that develop different traits over time (homologous structures); leads to speciation
convergent evolution: the process by which organisms that are not closely related evolve similar traits or behaviors to adapt to similar environments (analogous structures)
Causes of microevolution:
genetic drift
gene flow
nonrandom mating
mutations
natural selection
CHI-SQUARED TEST
chi-squared formula: determines at what point too much is “too much”
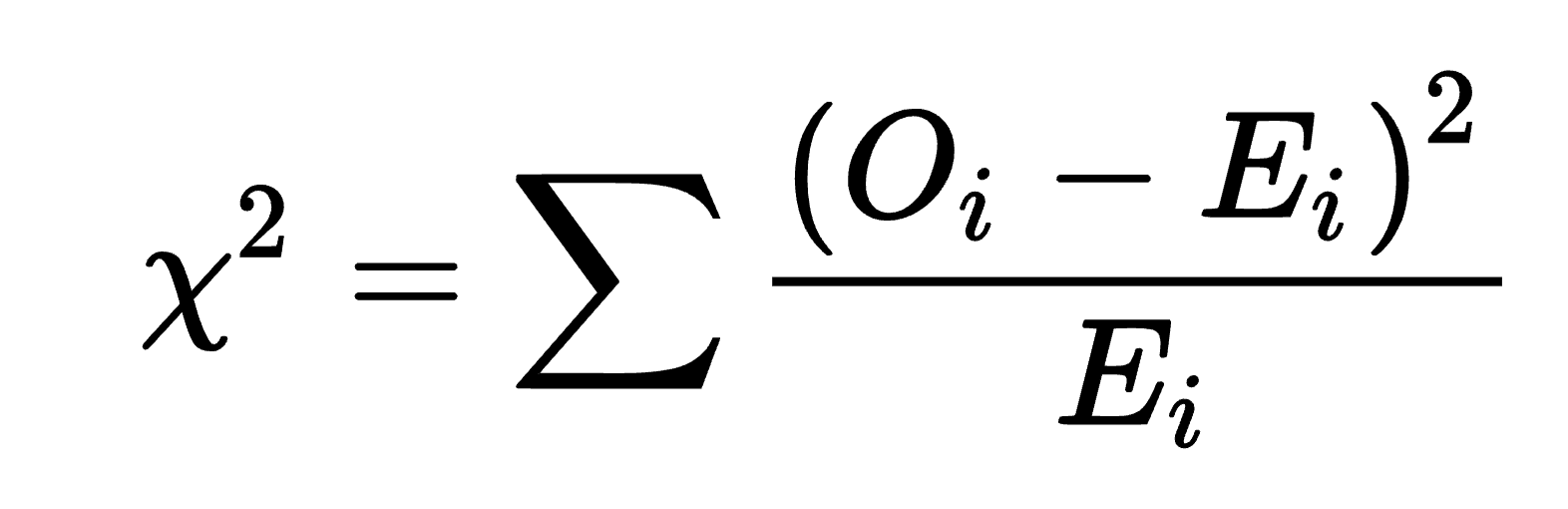
o = observed value; what you see (must be a whole number)
e = expected value; depends on the null hypothesis (can be a decimal)
x2= chi-squared value
null hypothesis: a general default position; assumes that there is no relationship between the 2+ phenomena.
steps for chi-squared:
predict (create null hypothesis)
collect data
calculate x2 for each group and add it together for the x2 sum
find the degrees of freedom (# of phenomena -1; n-1)
compare x2 sum to the critical value @ p=0.05
if x2 sum is:
greater than CV: result is insignificant (variation too large)
less than CV: result is significant (acceptable variation)
HARDY-WEINBERG EQUILIBRIUM
hardy-weinberg theorem:
allele frequencies: p+q=1
genotype frequencies: p2+2pq+q2=1
p and q are the frequencies of each allele (not quantities). they’re treated like percentages.
allele frequencies: p and q numbers
genotype frequencies: p2, 2pq, and q2 numbers
Hardy-Weinberg equilibrium: the population must be
very large
have random mating (no sexual selection/assortative mating)
no net mutation
no immigration/emigration
no natural selection
OVERALL: no evolution…this situation is like a “snapshot in time”
steps for h-w:
recessive phenotype: q2
always start with q
this is because p is dominant and gets mixed up with the heterozygous phenotypes
then, 1-q=p and proceed
METHODS OF SPECIATION
allopatric speciation: when a biological population becomes isolated from the rest of its species, preventing gene flow and leading to the development of new species; speciation in different places
isolation + time = potential for speciation
sympatric speciation: the split of a species into 2 reproductively isolated groups without geographical change; speciation in the same place
mostly in plants
autopolyploidy
allopolyploidy
reproductive isolation: [pre-zygotic] geographic, habitat/ecological, temporal, behavioral, mechanical, prevention of gamete fusion; [post-zygotic] hybrid invariability, infertility
geographic isolation: species are separated by physical geography and physically can’t reproduce with each other
habitat/ecological isolation: species are in the same area but in different habitats (ground vs. tree frog)
temporal isolation: species that have different mating cycles/timing
behavioral isolation: species have different mating rituals
mechanical isolation: species have morphological differences (parts don’t line up lol)
prevention of gamete fusion: gametes of one species function poorly with the gametes of another species/within the reproductive tract of another species
hybrid invariability: hybrid adults don’t survive in nature
infertility: hybrid adults are sterile or have reduced fertility
hybrid vigor: the hybrid is “best of both worlds”
gradualism: the theory that evolution occurs slowly and steadily over long periods of time (accumulation of small changes)
punctuated equilibrium: evolution happens in small bursts of rapid change separated by long periods of relative stability with little changes within a species