DNA Regulation
1) Differentiate between the structure of prokaryotic, eukaryotic, and viral DNA.
Prokaryotic DNA is found in a single circular chromosome,
- And sometimes they also have ==plasmids==, which are smaller circular pieces of DNA.
Eukaryotic DNA is linear and found in the nucleus, wrapped around proteins called ==histones==
Viral DNA can take various forms, but is usually linear, and is housed in a protein shell called a capsid. It can either be DNA or RNA
2) Explain how gene expression is controlled in eukaryotic and prokaryotic cells.
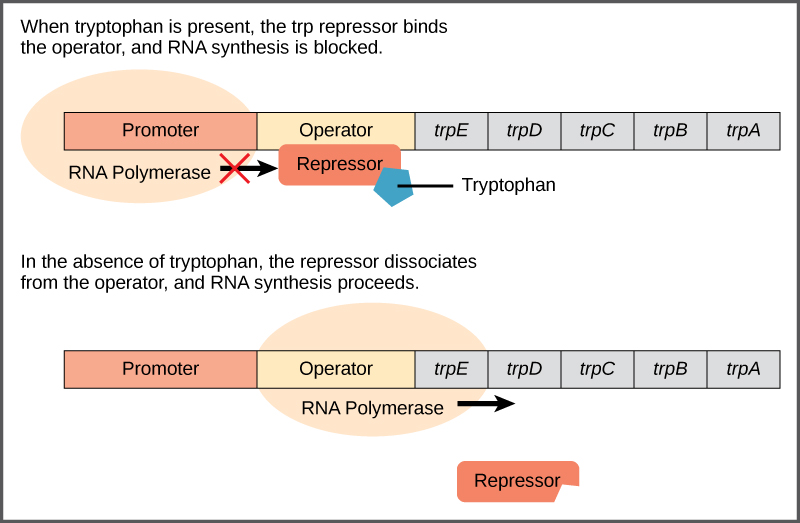
In prokaryotes, gene expression is controlled by ==operons==, which are groups of gene containing a single ==promoter== and ==operator==. The promoter is a site where RNA polymerase can bind to begin transcription. The operator is a site where a repressor can bind to prevent transcription.
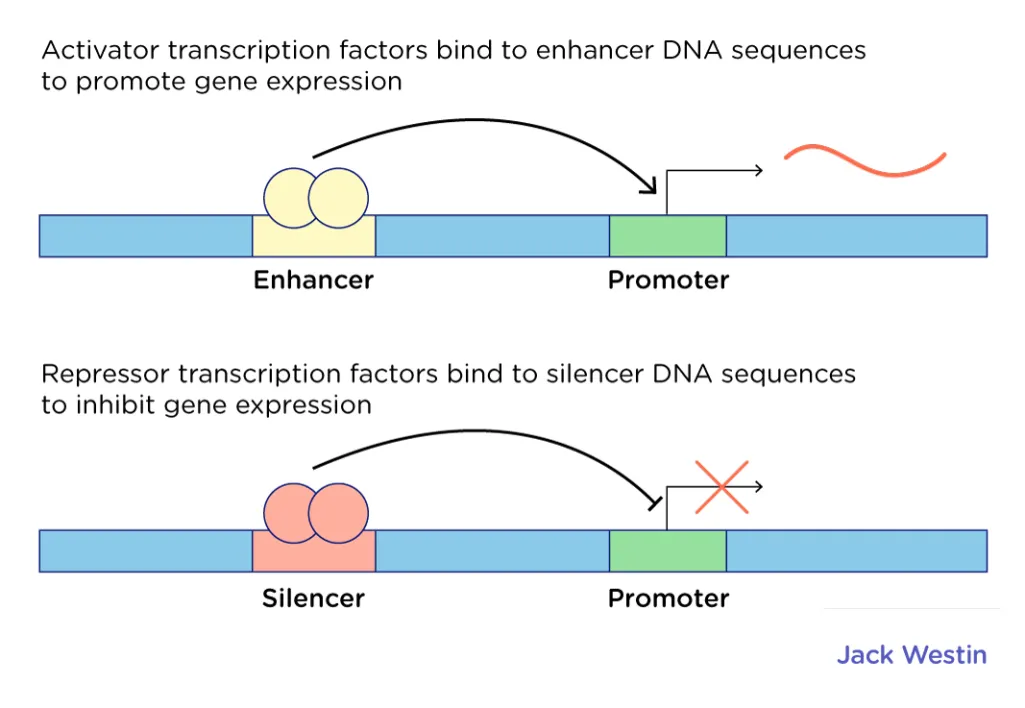
In eukaryotes, gene expression is controlled in the ==regulatory region==.
- This region has two parts, the ==enhancer region== and the ==silencer region.==
- To turn the gene on, ==activator transcription factors== bind to the enhancer region
- To turn the gene off, ==repressor transcription factors== bind to the silencer region
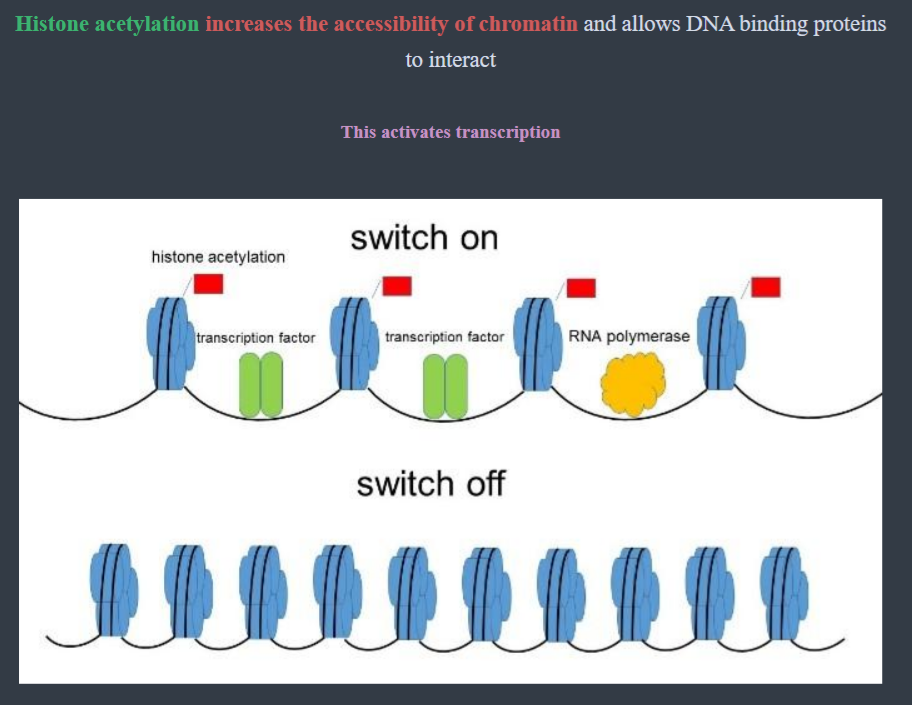
Eukaryotes can also regulate gene expression via the way DNA is wrapped around the histones
- If the DNA does not want to be read, then a methyl group is added in ==DNA Methylation== which causes the DNA to coil around the histone, making the gene unreadable (off)
- If the DNA does want to be read, then an acetyl group is added to the histone in ==Histone Acetylation== causing the DNA to unwind, making the gene accessible (on)
Vocabulary:
- Prokaryotes: simple, single-celled organisms with no nucleus
- Plasmids: small, circular pieces of DNA found in prokaryotic cells
- Transformation: process by which a prokaryotic cell takes up foreign DNA from its environment
- Recombinant DNA: DNA that has been manipulated to contain genetic material from different sources
- Restriction enzymes: enzymes that cut DNA at specific sites
- Operons: groups of genes in prokaryotes that are controlled by a single promoter and operator
- Promoter: site where RNA polymerase can bind to begin transcription
- Operator: site in an operon where a repressor protein can bind to prevent transcription
- Repressor: a protein that binds to an operator to prevent transcription
- Exons: coding regions of a gene that are transcribed into mRNA
- Introns: non-coding regions of a gene that are spliced out of the mRNA before translation
- Histones: proteins that DNA wraps around eukaryotes
- Histone acetylation: addition of acetyl group to histone causing DNA to unwind (gene on)
- DNA methylation: addition of methyl group causing DNA to coil (gene off)
- Control elements: regions of DNA that control gene expression
- Enhancer regions: regions of DNA that increase gene expression
- Silencer regions: regions of DNA that decrease gene expression
- Activator TF: a transcription factor that increases gene expression (binds to ==enhancer==)
- Repressor TF: a transcription factor that decreases gene expression (binds to ==silencer==)
- Lytic cycle: virus builds copies in cell and then bursts out
- Lysogenic cycle: virus integrates viral DNA into host DNA → eventually becomes lytic cycle
- Capsid: the protein shell of a virus
- Transduction: the transfer of genetic material using a virus as a vector